Human Liver Mononuclear Cells
Cat.No.: CSC-C9353W
Species: Human
Source: Liver
Morphology: Round
Cell Type: Mononuclear Cell
- Specification
- Background
- Scientific Data
- Q & A
- Customer Review
Human liver mononuclear cells (HLMCs) are a large group of immune cells in the liver that represent a wide variety of cell types. These cells are critical not only to maintaining normal liver function, but also to actively engage in immune responses and pathology. HLMCs are primarily composed of Kupffer cells, monocyte-derived macrophages (MDMs), liver sinusoidal endothelial cells (LSECs), liver epithelial progenitor cells, hepatic stellate cells, lymphocytes including T and B cells, and natural killer (NK) cells, among others.
Kupffer cells are the principal resident macrophages in the liver, responsible for clearing pathogens and damaged red blood cells from circulation, maintaining immune tolerance, and detecting tissue injury. MDMs are recruited and stimulated in response to inflammatory signals in the liver and contribute to inflammation. LSECs cooperate with Kupffer cells and hepatocytes, engaging in liver immune mechanisms and drug trafficking. These liver epithelial precursor cells can then transform into hepatocytes and bile duct cells. Hepatic stellate cells are activated during liver fibrosis and contribute to the accumulation of the extracellular matrix. Antibody production relies on lymphocytes. NK cells can even kill certain tumor cells and virus-infected cells directly, which is an important aspect of immune surveillance. Through collective function, these various HLMCs promote hepatic immune balance, regulate metabolism, and engage in many pathological processes. Thus, HLMCs are widely used in the study of how both innate and adaptive immune systems work, how diseases arise, and how drugs alter the immune system.
Expression of KIRs in PB and Liver NK, NKT and T Cells
The immune response to pathogens, alloantigens, and tumors is typically studied in peripheral blood, with limited information on the mechanisms in peripheral tissues such as the liver. Liver-specific immune responses involve various non-parenchymal cells and liver-associated lymphocytes, which differ phenotypically and functionally from peripheral blood mononuclear cells (PBMCs). Podhorzer et al. leverages liver perfusion methodologies to obtain LMCs, aiming for an in-depth analysis of natural killer (NK) and T cells (NKT) subsets to advance researchers understanding of liver-specific immune responses and their roles in liver diseases.
They compared the expression of KIR genes across peripheral blood, liver eluates, and a few mononuclear cell samples from liver biopsies, focusing on different KIRs due to limited sample sizes. In PB, KIR gene expression followed the order: CD56dim > NKT > CD56bright > T cells. In the liver, the order was: CD56dim > NKT > T > CD56bright cells. In PB, CD56bright NK cells had less than 10% KIR expression. However, CD56bright NK cells from the liver showed increased KIR3DL2 and KIR2DL3. CD56dim cells had higher KIR expression, with PB CD56dim cells showing significant levels of KIR2DS4. Comparative analysis revealed higher KIR2DL3 and KIR2DS1 expression in liver NKT cells compared to PB. For T cells, liver T cells exhibited significantly increased KIR expression: KIR2DL3, KIR3DL2, KIR2DS1, KIR2DS3, and KIR2DS4. Trends toward significance were also observed for KIR2DL2/2DS2, highlighting that three of the elevated KIR genes in liver T cells are activators, with KIR2DS4 and the weaker inhibitor KIR2DL3 showing the highest expression levels.
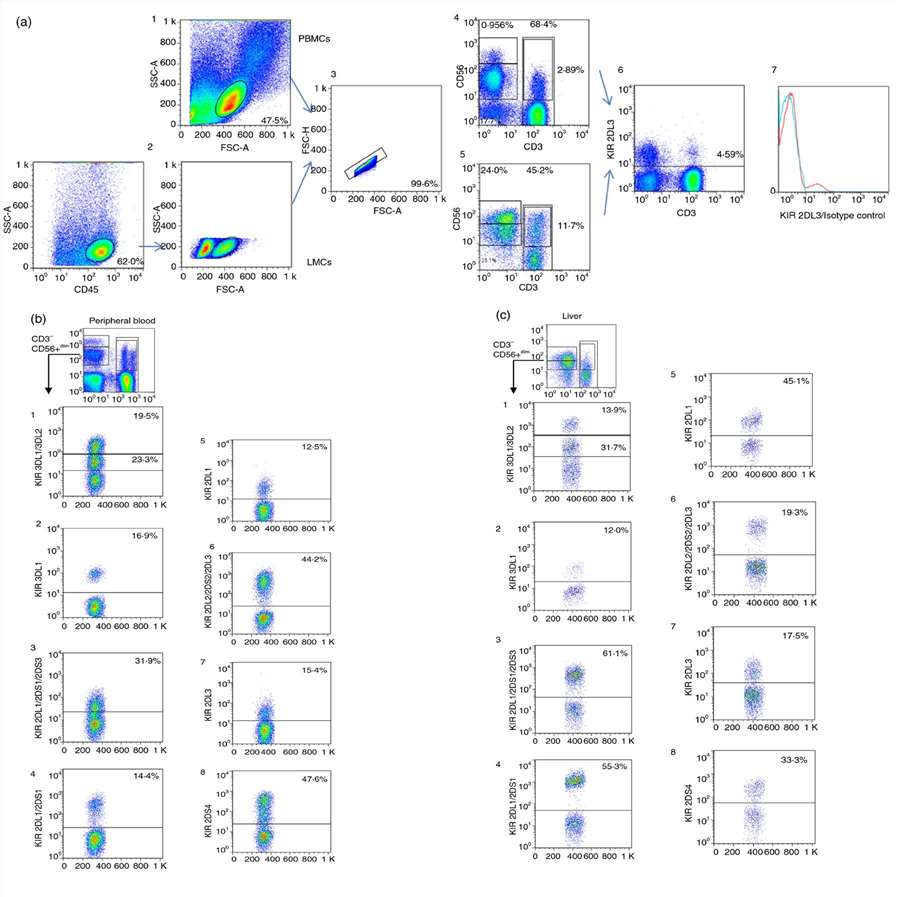 Fig. 1. Gating strategy for analyzing KIR expression in peripheral blood and liver CD45+ cells: comparison of CD56bright, CD56dim, NKT, and T Cell populations (Podhorzer A, Machicote A, et al., 2018).
Fig. 1. Gating strategy for analyzing KIR expression in peripheral blood and liver CD45+ cells: comparison of CD56bright, CD56dim, NKT, and T Cell populations (Podhorzer A, Machicote A, et al., 2018).
Innate Immunity Gene RNA Expression in Various Primary Liver Cells
The multifaceted liver not only processes metabolic information, detoxifies and digests food, it is also an important immune system. It contains the cell types that allow for sensing and targeting pathogens through a network of pattern recognition receptors (PRRs), including Toll-like receptors (TLRs) and RIG-I-like receptors. Faure-Dupuy et al. aimed to quantify expression and function of innate immune pathways in primary human liver cells for a better understanding of liver immune responses.
Primitive human liver hepatocytes (PHH), liver sinusoidal endothelial cells (LSEC), hepatic stellate cells (HSC) and Kupffer cells (KC) were isolated from several liver resections. On these unstimulated cells, baseline levels of 53 mRNAs involved in pathogen recognition and immune surveillance were identified via microfluidic high-throughput quantitative RT-qPCR. As controls, they used total peripheral blood mononuclear cells (PBMCs) and total nonparenchymal liver mononuclear cells (LMNCs) from different donors free of HBV, HCV and HIV. Comparatively, resident LMNCs exhibited higher gene expression levels at the basal state than PBMCs, highlighting the liver's crucial role in host immunity. Specifically, liver cells demonstrated elevated expression of several cytosolic DNA (DDX3, Ku70, DHX9, DHX36) and RNA sensors (RLR, RIG-I, MDA-5). TLR2 and TLR4 were notably elevated in KCs, while other TLRs had lower expression. Adaptors for TLR signaling (MyD88, TRIF, TRAM) and transcription factors (IRF1, 3, 7) had high basal expression. NOD1 and NOD2 were minimally detected, but their adaptor RIP2 was present. Inflammasome receptor mRNAs were low, though ASC and caspase 1 proteins were detectable.
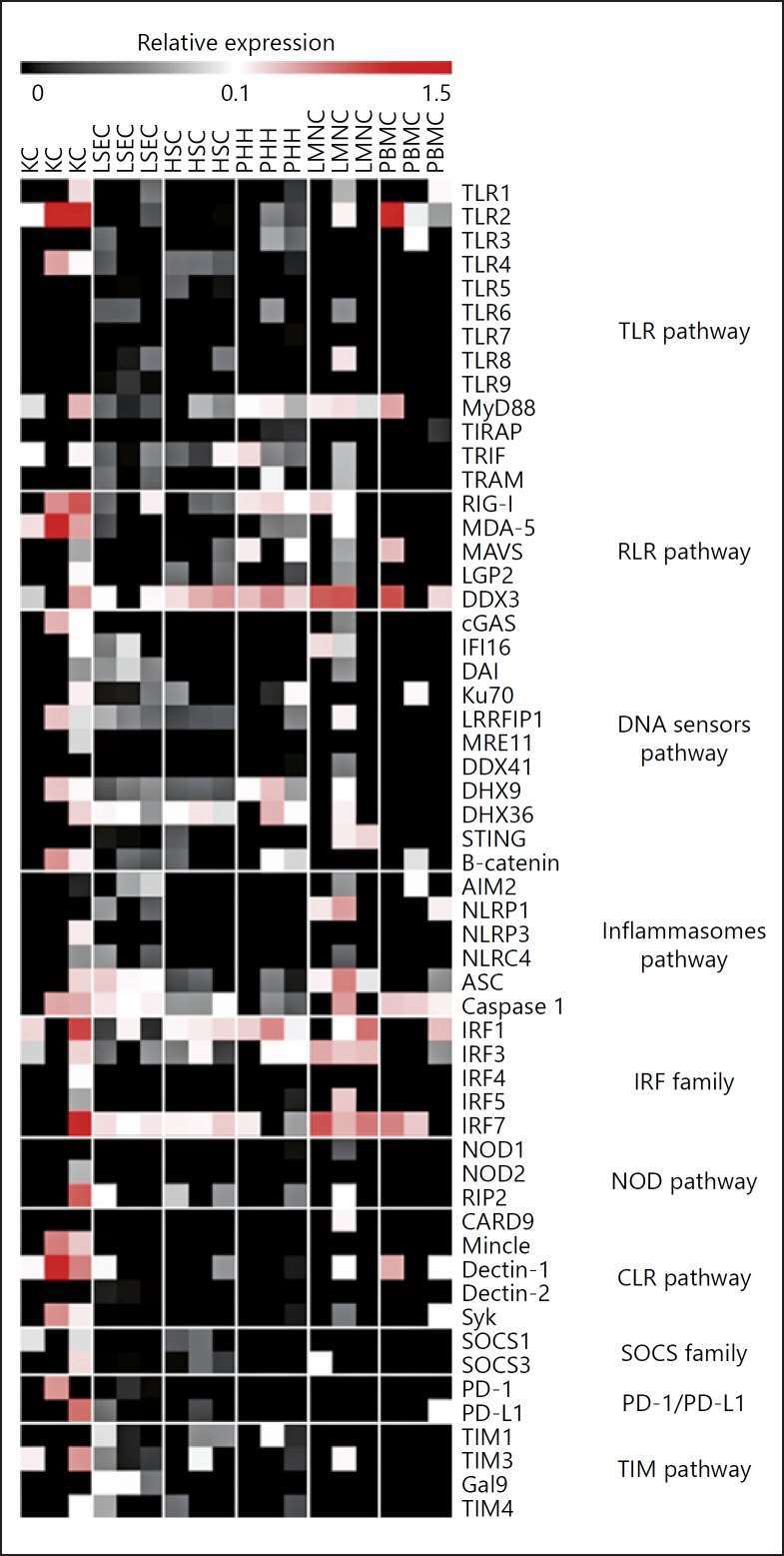 Fig. 2. Liver cells expression of innate sensors and related molecules in PHH, LSEC, HSC, KC, LMNC, and PBMC (Faure-Dupuy S, Vegna S, et al., 2018).
Fig. 2. Liver cells expression of innate sensors and related molecules in PHH, LSEC, HSC, KC, LMNC, and PBMC (Faure-Dupuy S, Vegna S, et al., 2018).
Ask a Question
Write your own review