FISH-Based Clonality Analysis Services
- Service Details
- Workflow
- Features
- Case Studies
- FAQ
- Explore Other Options
A well-characterized, clonally derived cell line is a regulatory expectation and a cornerstone of consistent biopharmaceutical manufacturing, especially for Investigational New Drug (IND) submissions. Demonstrating clonality helps minimize cell bank heterogeneity and supports product safety, quality, and long-term manufacturing consistency. Regulatory agencies, including the FDA and EMA, require evidence that production cell lines originate from a single progenitor cell as part of an overall control strategy.
Technologies such as single-cell sorting and imaging can provide supporting evidence, but re-derivation through additional limiting dilution is often time-consuming, costly, and may compromise established growth or productivity profiles. This poses challenges under accelerated development timelines. Creative Bioarray's FISH-based clonality analysis services provide robust, visual, and regulator-aligned evidence to support clonal characterization without the need for re-cloning, helping clients advance programs with confidence and efficiency.
Why Choose FISH for Clonality Analysis?
| Flow Cytometry | PCR-based Assays | NGS | FISH (Our Approach) | |
| Level of Resolution | Protein/ phenotype level only | Locus-specific | Base to genome scale | Single-cell, spatial, chromosome-level |
| Detection of Structural Variants | ✕ | Limited | ✓ (dependent on depth) | ✓ Direct visualization of CNV, translocations, amplifications |
| Single-Cell Clonality Readout | ✕(population-based) | ✕ | Partial(computational inference) | ✓ True cell-by-cell assessment |
| Low-Frequency Clone Detection | Limited sensitivity | Risk of allele dropout | Requires deep sequencing | ✓ Visual enumeration, robust for rare clones |
| Cost & Turnaround | Low | Low | Medium–High | Medium, fast for known targets |
Our Clonality Analysis Capabilities
Lifecycle-based clonality assessment
Our FISH-based clonality analysis service provides a high-confidence assessment of the genomic stability of Master Cell Banks (MCB), Working Cell Banks (WCB), and End-of-Production Cells (EOPC).
Using advanced multi-color FISH platforms, our experts evaluate:
- Chromosome number and ploidy status
- Structural abnormalities (translocations, deletions, duplications, amplifications)
- Clonal uniformity and population heterogeneity
- Genomic stability across the entire production lifecycle
Each project is supported by a fully traceable workflow, including standardized sample handling, optimized hybridization chemistry, automated imaging, and expert cytogenetic review.
You will receive a regulator-ready report detailing karyotype composition, transgene integration pattern, and cell line clonality discussion.
Cell line-specific clonality assessment
Our FISH-based clonality analysis services are designed to accommodate a wide range of cell lines, including the industry-leading Chinese Hamster Ovary (CHO) and Human Embryonic Kidney (HEK293) cell lines. For each cell line, we provide:

- Customized chromosome preparation and metaphase optimization
- Probe design and synthesis tailored to your target loci
- High-resolution metaphase FISH analysis
- Clone-level interpretation with quantitative cell-by-cell assessment
Whether you are establishing a new production clone or monitoring long-term stability, our team provides genomic insights that enhance cell line characterization and support robust, reliable manufacturing workflows.
FISH-Based Clonality Analysis Workflow
Define Project Scope
Identify sample stage (MCB/WCB/EOPC), targets, and analysis depth.
Prepare Chromosomes & FISH Probes
Generate high-quality metaphase slides and custom-labeled probes.
FISH Hybridization & Imaging
Multi-color FISH with automated imaging captures consistent, high-resolution data.
Analyze & Report
Expert review delivers traceable, regulator-ready reports detailing clonal structure and genomic stability to support decision-making.
Key Features of Our FISH-Based Clonality Analysis
Cell-level certainty
Identify every integration site and visualize true clonality without inference or assumptions.
Deep sampling
100–200 cells per assay reveal heterogeneity that bulk genomic methods routinely overlook.
Rapid, decision-ready results
4-6 weeks delivery with expert annotation supporting clone selection and stability decisions.
Proven sensitivity
Detect rare subclones and integration-site variation beyond the limits of PCR and short-read NGS.
Budget-friendly options
Designed for screening, comparability, and regulatory-aligned documentation needs.
Case Studies
Case 1: FISH-Based Clonality Analysis for MCB
Chromosomal rearrangements (or genetic plasticity) would occur in CHO cells over a 30-day period of culture, which is the case with an MCB. Out of the 200 cells that were randomly captured and analyzed, 200 cells (100%) showed uniform single-site integration in the centromeric region of Chr.10.
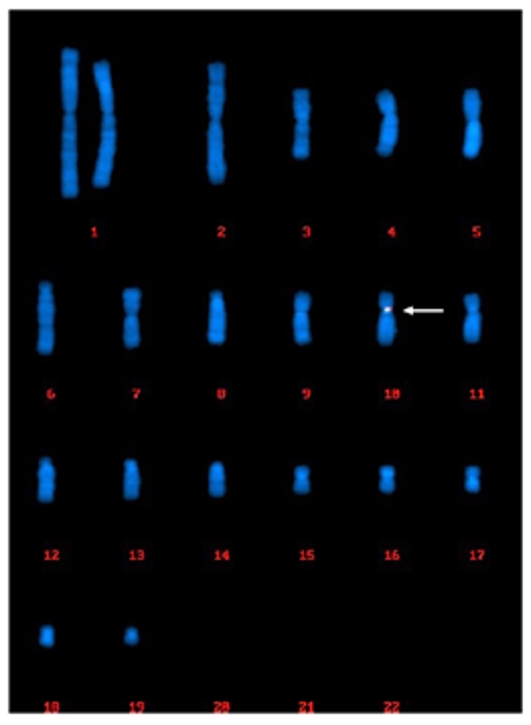 Fig. 1 (200/200 cells) Centromeric region of chr.10.
Fig. 1 (200/200 cells) Centromeric region of chr.10.
Case 2: FISH-Based Clonality Analysis for F7C2-P3C63 Cell Line
In 95% of the cell population analyzed, we have identified a dominant pattern characterized by the presence of FISH signals on two chromosomes, which is the long arm of the large submetacentric chromosome, positioned at #1 on the karyograms and the long arm of the medium-sized submetacentric chromosome, positioned at #6 on the karyograms (95 out of 100 cells). Apart from the dominant pattern, another two patterns were observed. In four cells, FISH signals were observed only on chromosome 1. In one cell, FISH signals were observed on chromosome 6 and an unknown chromosome.
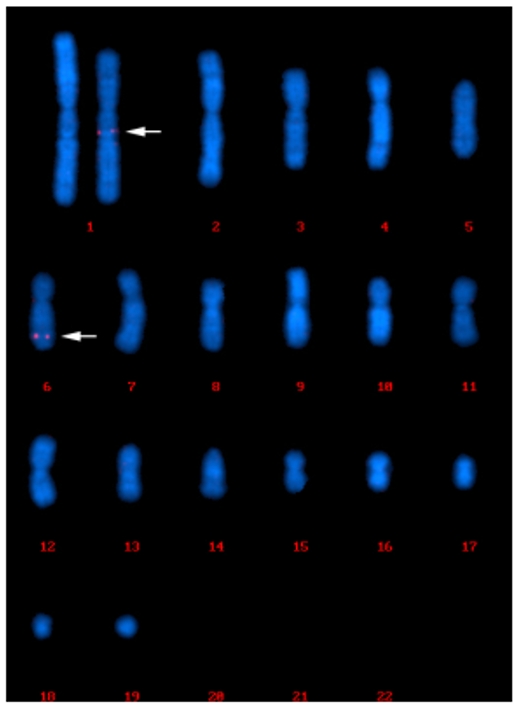 Fig. 2 FISH signals were observed on chromosome 1 and chromosome 6.
Fig. 2 FISH signals were observed on chromosome 1 and chromosome 6.
FAQ
1. How does FISH-based clonality analysis work?
FISH-based clonality analysis works by hybridizing fluorescently labeled DNA probes to specific target sequences within the cells' chromosomes. By visualizing these probes under a fluorescence microscope, researchers can identify and quantify the presence of specific genetic markers. Consistent patterns across multiple cells indicate clonality, whereas heterogeneous patterns suggest a polyclonal or mixed-cell population.
2. What are the advantages of using FISH for clonality analysis compared to other methods?
The advantages of using FISH for clonality analysis include high specificity and sensitivity for detecting genetic markers, the ability to visualize genetic changes at the single-cell level, relatively rapid and straightforward procedure compared to molecular techniques like PCR or next-generation sequencing, the capability to detect both large chromosomal abnormalities and smaller genetic changes.
3. What are the applications of FISH-based clonality analysis?
Confirming the monoclonality of cell lines used in research and biopharmaceutical production; assessing the clonal evolution of cancer cells during treatment; and ensuring the genetic uniformity of genetically engineered cell lines for therapeutic purposes.
At Creative Bioarray, our FISH-based clonality analysis services are designed to meet the stringent requirements of regulatory bodies, such as the FDA and EMA, ensuring that your data is aligned with the necessary guidelines. If you have any special needs or questions regarding our services, please feel free to contact us or make an online inquiry.
References
- Frye, Christopher, et al. "Industry view on the relative importance of "clonality" of biopharmaceutical-producing cell lines." Biologicals 44.2 (2016): 117-122.
- Wurm, Florian M., and Maria João Wurm. "Cloning of CHO cells, productivity, and genetic stability-a discussion." Processes 5.2 (2017): 20.
- ICH Q5D Derivation and characterization of cell substrates used for the production of biotechnological/biological products (CPMP/ICH/294/95).
- Welch, J. (2017). Tilting at clones: A regulatory perspective on the importance of "Clonality" of mammalian cell banks. CDER/OPQ/OBP/DBRRIV April 24, 2017.
Explore Other Options