Key Techniques in Primary, Immortalized and Stable Cell Line Development
Primary Cell Lines
Primary cell isolation techniques are employed to extract original, non-passaged cells from tissues or organs for subsequent culture and characterization. Commonly utilized methods include suspension centrifugation, direct explant method, enzymatic digestion, and mechanical dissociation.
Suspension centrifugation is primarily applied to tissue samples derived from blood, pleural effusions, ascites, and organ perfusate. For tissue blocks, cells in the tissue can be ripped apart using mechanical dissociation or enzymatic digestion. Enzymes for this include collagenase, Dispase, hyaluronidase, and trypsin. In addition, chemicals are also implicated in weakening cell-cell and cell-ECM connectivity to isolate primary cells. Agents like ethylenediaminetetraacetic acid (EDTA) and EGTA are often used to reverse ion-dependent bonding between cells and ECM by chelating cations.
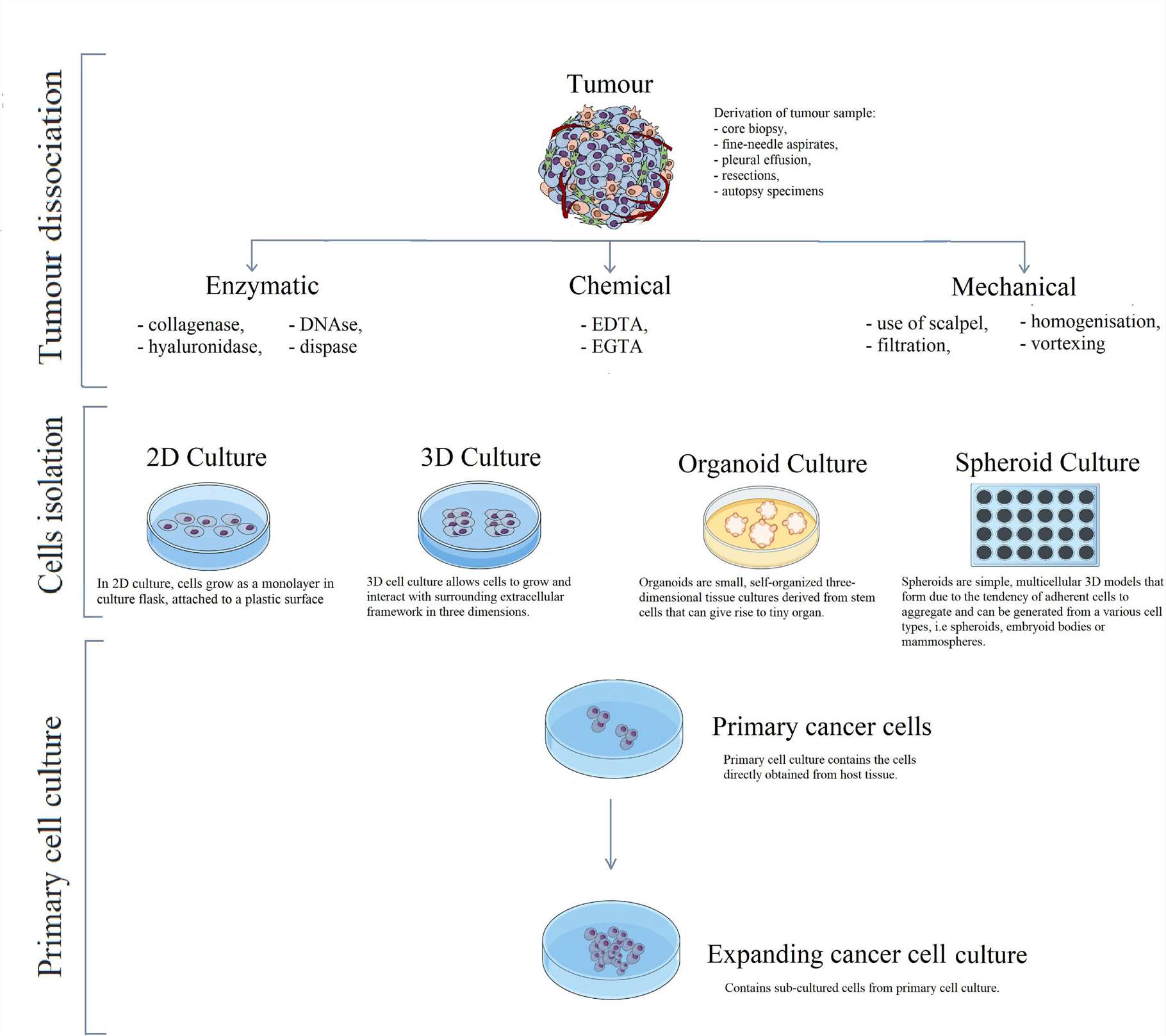 Fig. 1. Schematic visualisation of primary cell line development, taking cancer cells as an example (Richter M, Piwocka O, et al., 2021).
Fig. 1. Schematic visualisation of primary cell line development, taking cancer cells as an example (Richter M, Piwocka O, et al., 2021).
Single-cell isolation technology
Isolation of individual cells is a major challenge in medical and biological research. There are two kinds of conventional cell-separation methods: labeled and label-free. Labeled techniques use cell specific antibodies to separate (such as Fluorescence-Activated Cell Sorting (FACS) and Magnetic Activated Cell Sorting (MACS), whereas label-free approaches are based on cell biology. This selective separation of target cells from mixed molecules is essential to therapy, diagnosis and research. The following are standard single-cell isolation technologies:
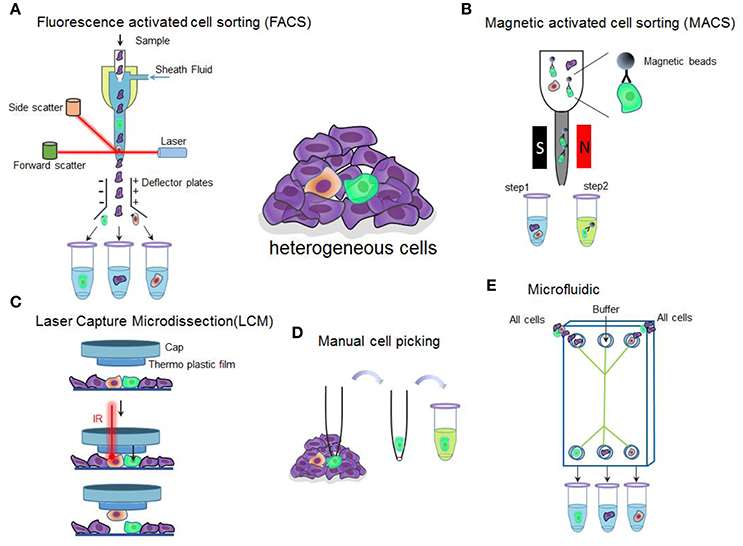 Fig. 2. Overview of single-cell isolation technologies (Hu P, Zhang W, et al., 2016).
Fig. 2. Overview of single-cell isolation technologies (Hu P, Zhang W, et al., 2016).
- Flow cytometry: Flow cytometry combines laser stimulation and fluorescent staining to sort and label cells. The FACS technique isolates individual cells.
- Magnetic activated cell sorting (MACS): MACS is a non-invasive cell-cluster splitting method that uses magnetically imprinted antibodies. Polarised-labelled cells are suspended in a magnetic field, and target cells can be isolated when the magnetic field is eliminated.
- Laser capture microdissection (LCM): LCM is used to remove individual cells or cell fragments from a solid tissue. It encompasses contact-based extraction, contact-free gravity-assisted cutting and laser pressure catapulting. This method is primarily used to study cellular architecture and pathophysiological processes in heterogeneous tissues, particularly in solid tumor research.
- Limiting dilution: Limiting dilution involves diluting a cell suspension manually or using robotic pipettes, allocating them into small-volume wells with the expectation of having a single cell per well. Due to the statistical nature of the method, the actual efficiency is low, and additional microscopic confirmation is required.
- Manual cell picking: With the aid of a microscope and micromanipulator, workers can remove individual cells and pipette them into collection tubes. This method is used for cell suspension, bacterial analysis, reproductive medicine, and other applications.
- Microfluidics: Microfluidic technologies use microchannels for single-cell analysis and processing such as droplet isolation, pneumatic membrane valves, and hydrodynamic traps. This allows sample preprocessing to be detected on the microfluidic chip and has several advantages such as low price, high performance and small sample size.
- Acoustic flow cytometry: Acoustic flow cytometry is an emerging technology that utilizes sound waves and microfluidics to control fluids and particles at the microscopic level. Acoustic flow is the perfect method to remove cells and living material from their environment: it is non-labelled, non-touchable and biocompatible.
- Dielectrophoresis-Based Cell Separation: This technique isolates cells by exploiting the diverse dielectric nature of cells in a chaotic electric field. This method can efficiently isolate cancer cells with an efficiency rate exceeding 95%.
- Biochemical cell separation:
Immunoaffinity methods utilize specific antibodies or aptamers to capture cells, serving as a common separation strategy, particularly in tumor cell isolation. Furthermore, synthetic chemical antibodies—aptamers—have been developed to effectively and specifically capture target cells.Cell adhesion methods achieve separation by enhancing the interaction between target cells and channel surfaces, making it one of the few techniques that do not rely on labels or size. This approach is particularly suitable for isolating certain components from blood samples.
Table. 1. Overview of single cell isolation techniques (Hu P, Zhang W, et al., 2016).
| Techniques | Throughput | Advantage | Disadvantage |
| Fluorescence-activated cell sorting (FACS) | High | High specificity multiple parameters | Large amount of material,dissociated cells, high skill needed |
| Magnetic-activated cell sorting (MACS) | High | High specificity, cost effective | Dissociated cells, non-specific cell capture |
| Laser capture microdissection (LCM) | Low | Intact fixed and live tissue | Contaminated by neighboring cells, high skill needed |
| Manual cell picking | Low | Intact live tissue | High skill needed, low throughput |
| Microfluidic | High | Low sample consumption, integrated with amplification | Dissociated cells, high skill needed |
Immortalized Cell Lines
What is cell immortalization?
Cell immortalisation is a process where cells can (through genetic mutation or triggers) avoid senescence and death that would normally be caused by serial passaging and continue to multiply forever in vitro. Other than stem cells and germ cells, most eukaryotic cells escape growth arrest phases (M1 and M2) in certain circumstances by triggering telomerase, keeping telomeres intact, and avoiding senescence and allowing uncontrolled cell division.
Why immortalize cells?
1) Immortalized cells are able to proliferate continuously, breaking through the limitations of traditional cells that are less transmissible and slower to proliferate, providing a large number of cell resources. Thereby shortening the experimental cycle and reducing the cost of isolating primary cells.
2) Due to the similarity in behavior between immortalized and tumor cells, they are a key tool for understanding the mechanisms of tumorigenesis and developing therapies to inhibit cancer cell proliferation.
3) Immortalized cell lines are genetically consistent and can create stable cell banks, which is essential for obtaining consistent and reproducible experimental results.
4) They are ideal models for studying biological processes such as cell proliferation, differentiation, apoptosis and senescence.
Table. 2. Comparison of primary and immortalised cell lines (Richter M, Piwocka O, et al., 2021).
| Feature | Primary cells | Immortalised cell lines |
| Biological relevance | Higher | Lower |
| Lifespan and proliferation | Limited | Unlimited |
| Molecular properties | Preserve characteristic cells' behaviours and cross-talk between cells | Untypical cellular behaviours may occur |
| Phenotype | High heterogeneity | Lower heterogeneity |
| Genetics | Original genome | Altered genome, accumulation ofgenetic aberrations |
| Relevance in vivo | Best experimental models for in vivo studies | Low relevance for in vivo studies |
| Handling | Needs optimised culture conditions and media | Established media from manufacturer |
| Ethics | Require Ethical Commission approval and patient consent | Do not require additional documents |
Methods of immortalized cells construction:
- Proto-oncogene or tumor suppressor gene mutations: Gene mutations can turn on proto-oncogenes or turn off tumor suppressor genes, thereby bypassing normal cell cycle checkpoints, cell death and senescence, and thereby allowing unconstrained cell proliferation.
- Telomerase activation: Telomerase generates and expands telomeres, which preserves chromosomal integrity during cell division, which contributes to continual cell growth.
- Chemical and radiation factors: Ionizing radiation and chemical carcinogens can induce immortal cell fate via chromosome mutations and gene-expression changes, but can lead to genomic instability.
- Viral infection:
Virus oncogenes interfere with tumour suppressor genes, allowing cells to grow unrestricted, bypassing senescence, and thus granting the potential for infinite growth.
Human Papillomavirus (HPV): HPV infection may make epithelial cells immortal.
Epstein-Barr Virus (EBV): EBV suffocates B lymphocytes, producing immortalized lines of lymphocytes.
Simian Virus 40 (SV40): SV40's T antigen modulates DNA replication and gene expression at the cell nucleus, leading to cell immortalisation.
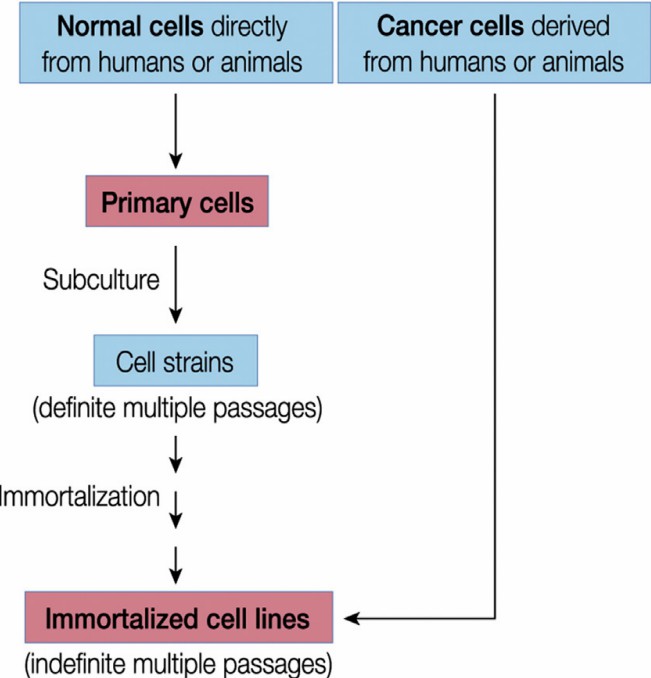 Fig. 3. A diagram depicting steps involved in preparation of primary cell versus immortalized cell line (Ryu W S, 2017).
Fig. 3. A diagram depicting steps involved in preparation of primary cell versus immortalized cell line (Ryu W S, 2017).
Stable Cell Lines
Stable transfected cell lines (also called stable expression cell lines) are engineered from a previously established cell line to continue overexpressing or suppressing a given gene. They are used widely in the production of recombinant proteins, signal transduction, drug target identification, and immunotherapeutics. Vector design, cell transfection, selection, culture expansion, and clone identification are part of the design.
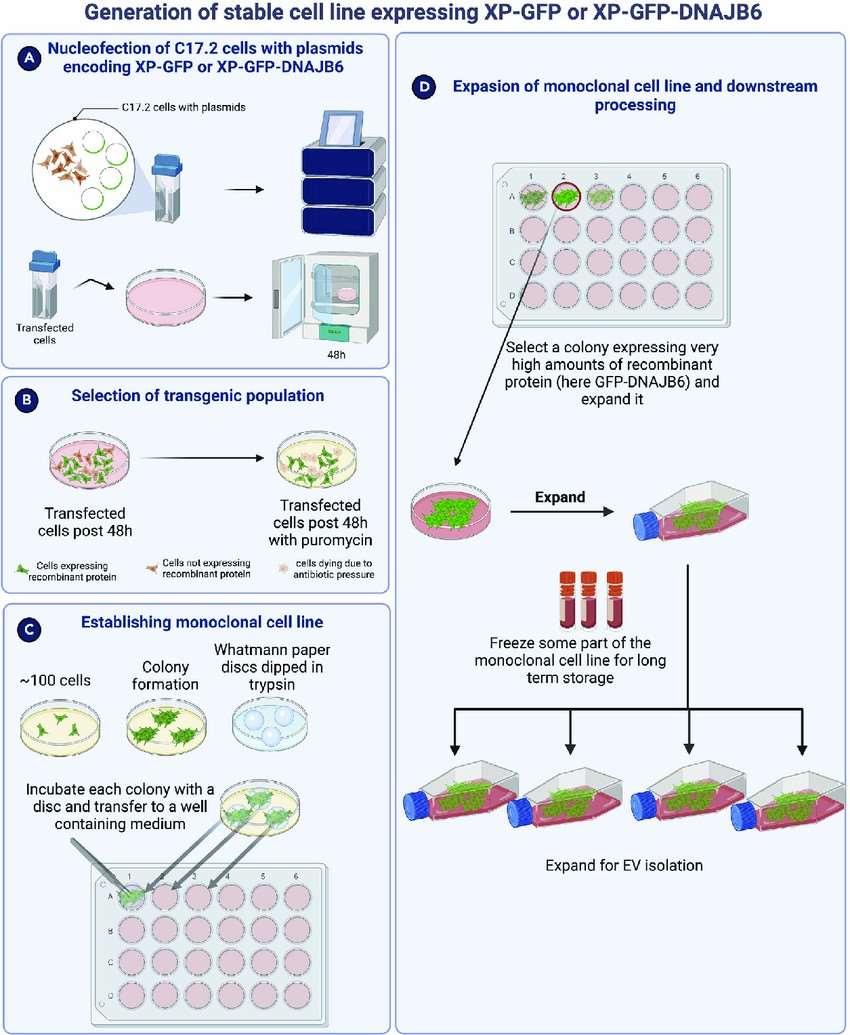 Fig. 4. Schematic overview of generation of monoclonal stable cell line expressing the recombinant protein of interest (Joshi BS, Zuhorn IS, et al., 2023).
Fig. 4. Schematic overview of generation of monoclonal stable cell line expressing the recombinant protein of interest (Joshi BS, Zuhorn IS, et al., 2023).
Methods of Stable Cell Lines Construction:
Lentivirus-mediated cell infection:
Lentiviral vectors are a classic way to generate stable cell lines. This technique allows you to deliver shRNA, overexpression constructs, CRISPR, reporter genes, and so on. Second- and third-generation lentiviral packaging systems are capable of generating high-titer lentivirus vectors. However, due to the packaging capacity limitations of lentiviruses, they are less suitable for large transcript regions, posing constraints on larger genes. Additionally, being a retrovirus, lentivirus-mediated gene integration can influence the translation of certain genes.
Plasmid-mediated cell transfection:
- Transposon systems: Transposons (or "jumping genes") make it possible to "cut and paste" pieces of DNA inside or between chromosomes. Engineered transposons include short terminal ITR sequences encircling the transposase code. The most well-known transposon systems are PiggyBac, Sleeping Beauty and Tol2.
- CRISPR knock-in systems: CRISPR/Cas9 supports targeted gene knock-in by providing exogenous templates (Donor vectors) that exploit the cell's endogenous repair systems including non-homologous end joining (NHEJ) and homology-directed repair (HDR).
Creative Bioarray Relevant Recommendations
| Products & Services | Description |
| Cell Immortalization Service | Reliable and risk-free cell immortalization from fibroblasts, epithelial cells, muscle cells from human, rat, mouse, porcine and bovine. |
References
- Ryu W S, Immortalised Cell Line. Molecular Virology of Human Pathogenic Viruses. 2017. 47-62
- Joshi BS, Zuhorn IS. Preparation of chaperone-loaded neural stem cell-derived extracellular vesicles to reduce protein aggregation in Huntington's disease cellular models. STAR Protoc. 2023. 4(1):102134.
- Richter M, Piwocka O, et al. From Donor to the Lab: A Fascinating Journey of Primary Cell Lines. Front Cell Dev Biol. 2021. 9:711381.
- Hu P, Zhang W, et al. Single Cell Isolation and Analysis. Front Cell Dev Biol. 2016. 4:116.