Classification, Isolation Techniques and Characterization of Exosomes
Exosomes are small membrane-bound vesicles secreted by various cell types, including both prokaryotic and eukaryotic cells. These extracellular vesicles play a crucial role in intercellular communication and are involved in numerous physiological and pathological processes.
Introduction
Exosomes were initially discovered in the 1980s as small vesicles secreted by reticulocytes during the process of maturation into red blood cells. Since then, research on exosomes has expanded, revealing their presence in various biological fluids, including blood, urine, saliva, and cerebrospinal fluid. Exosomes are released by both healthy and diseased cells, and their cargo, including proteins, nucleic acids, lipids, and metabolites, reflects the physiological or pathological state of the originating cells.
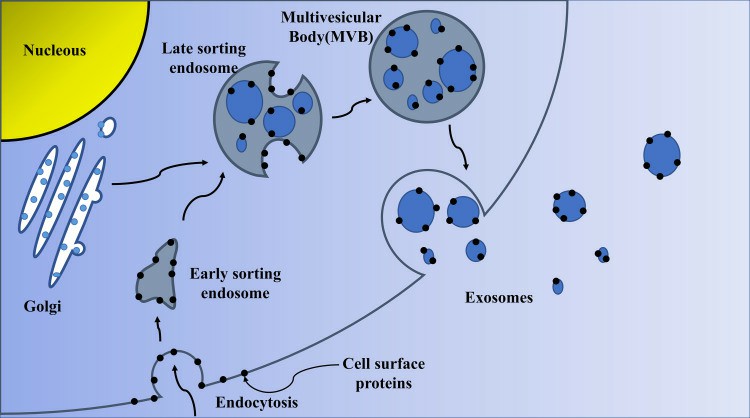 Fig. 1 Biogenesis of exosomes. (Zhang Y, et al., 2020)
Fig. 1 Biogenesis of exosomes. (Zhang Y, et al., 2020)
Exosomes are known to mediate cell-to-cell communication by transferring their cargo to recipient cells, thereby influencing cellular functions. They have been implicated in various biological processes, including immune response modulation, tissue regeneration, and cancer progression. Due to their unique properties and potential as biomarkers and therapeutic agents, exosomes have garnered significant attention in the field of biomedical research.
Classification
Exosomes are divided into natural exosomes and engineered exosomes based on whether they have been artificially modified. Ulteriorly, natural exosomes are divided into animal-derived exosomes and plant-derived exosomes. Because exosomes are produced under normal and tumor conditions, animal-derived exosomes are further divided into normal exosomes and tumor exosomes.
Isolation Techniques
Accurate isolation of exosomes is crucial for their study and downstream applications. Several methods have been developed for exosome isolation, each with its advantages and limitations. Common techniques include ultracentrifugation, density gradient centrifugation, precipitation methods, size exclusion chromatography, and immunoaffinity-based capture.
- Ultracentrifugation is considered the gold standard method for exosome isolation. It involves sequential centrifugation steps to pellet exosomes based on their density. However, ultracentrifugation can be time-consuming, requires specialized equipment, and may result in co-purification of other extracellular vesicles or contaminants.
- Density gradient centrifugation utilizes density gradients, often using iodixanol or sucrose, to separate exosomes from other particles based on their buoyant density. This technique provides higher purity but is also time-consuming and requires expertise.
- Precipitation methods, such as the use of polyethylene glycol (PEG) or commercial reagents, offer a more convenient and rapid approach to exosome isolation. These methods rely on the precipitation of exosomes from biological fluids, followed by centrifugation to obtain a pellet. However, they may co-precipitate contaminants and require additional purification steps.
- Size exclusion chromatography separates exosomes based on their size using columns packed with porous beads. This technique provides relatively high purity and is compatible with large-volume samples. However, it may not effectively separate exosomes from particles of similar size, such as lipoproteins.
- Immunoaffinity-based capture relies on the specific interaction between exosome surface markers and antibodies immobilized on solid supports. This technique allows for the isolation of exosomes expressing a particular marker, enabling the enrichment of specific subpopulations. However, it requires prior knowledge of the exosome surface markers and may result in the loss of non-targeted exosomes.
Characterization
In general research, we consider the identification of isolated exosomes from three levels, including electron microscopy identification of exosome morphology, NTA identification of exosome size, and Western Blot identification of exosome surface protein markers. Exosome characterization methods are mainly divided into two types: external characterization (mainly morphology and particle size detection) and inclusion characterization (like membrane protein, and lipid raft).
- Electron microscopy provides morphological information by visualizing exosomes under high-resolution microscopy. It allows for the assessment of the size, shape, and membrane integrity of exosomes. However, it does not provide detailed information about the cargo or molecular composition of exosomes.
- Nanoparticle tracking analysis (NTA) is a technique that utilizes laser light scattering to measure the size distribution and concentration of exosomes in a sample. It provides quantitative data on the size and concentration of exosomes and can be used to monitor changes in exosome populations under different conditions. However, NTA cannot distinguish between exosomes and other extracellular vesicles or particles of similar size.
- Flow cytometry is a potent technique for phenotypic assessment of exosomes. Through the application of particular antibodies or fluorescent probes to label exosomes, flow cytometry provides data on surface marker expression or specific protein presence. It permits the characterization of exosome subpopulations and the identification of exosomes originating from different cell types.
- Western blotting is a commonly used method to analyze the protein composition of exosomes. This method includes the separation of exosome proteins via gel electrophoresis, followed by transfer onto a membrane and detection with specific antibodies. The results of Western blotting can provide information about the presence or absence of specific proteins in exosomes and can be used to validate the expression of known exosome markers. However, it has constraints in identifying proteins with low concentrations and may not capture the entire proteome of exosomes.
Creative Bioarray Relevant Recommendations
- Creative Bioarray provides reliable techniques for exosome isolation from different sample matrices. We also provide exosome isolation using specified techniques if required by customers. We will perform QC assays to ensure the integrity and purity of the isolated materials.
- We provide comprehensive support for your exosome identification by including the morphology assay, purity, and quantity assay, particle size distribution analysis, and exosome-specific markers expression.
Reference
- Zhang Y, et al. (2020). "Exosome: A Review of Its Classification, Isolation Techniques, Storage, Diagnostic and Targeted Therapy Applications." Int J Nanomedicine. 15, 6917-6934.