Hepatoma Cells
- Background
- Applications
- Scientific Data
- FAQ
Hepatoma cells, particularly HepG2 cells, are a type of cell line derived from liver cancer. They are characterized by their unlimited lifespan, stable phenotype, high availability, and ease of handling, which makes them an invaluable tool in biological and medical research. However, one of their limitations is their lower expression of some metabolic activities compared to normal hepatocytes, or liver cells. Despite this, HepG2 cells, which originate from a human hepatoma (a type of liver tumor), are most commonly used in studies related to drug metabolism and hepatotoxicity. These cells retain many characteristics of normal hepatocytes, and thus, they provide a useful model for studying liver diseases, the liver's response to drugs, and other related topics. They are widely used in labs around the world for this purpose.
Biological Properties of Hepatoma Cells
- Cell morphology and growth characteristics. Hepatoma cells often exhibit a more irregular and disorganized appearance, with a less uniform shape and size compared to the well-organized structure of healthy hepatocytes. This morphological transformation is believed to be a reflection of the underlying genetic and molecular alterations that drive the malignant transformation of these cells.
- Proliferation and cell division patterns. In terms of growth patterns, hepatoma cells demonstrate a significantly higher proliferation rate than normal liver cells. These cells are characterized by a more aggressive cell division pattern, with a shorter doubling time and a greater capacity for self-renewal.
- Metabolic and functional differences from normal liver cells. Unlike normal liver cells, which primarily rely on oxidative phosphorylation for energy production, hepatoma cells exhibit a pronounced shift towards aerobic glycolysis, a phenomenon known as the "Warburg effect." This metabolic reprogramming provides these cancer cells with a competitive advantage, as it allows them to generate ATP more quickly, even in the presence of oxygen. Furthermore, hepatoma cells often display dysregulation of various liver-specific functions, such as bile acid synthesis, xenobiotic metabolism, and the production of essential proteins.
Drug Testing and Toxicology Studies
Hepatoma cells exhibit many of the characteristic features of liver cells, including drug-metabolizing enzymes and transporters, making them a relevant system for assessing the potential liver-related adverse effects of new therapeutics. Researchers can utilize hepatoma cells to screen for hepatotoxicity, study the metabolism and pharmacokinetics of drugs, and investigate the underlying mechanisms of liver injury, helping to identify and mitigate potential liver-related toxicities early in the drug development process.
Liver Disease and Regeneration Studies
Hepatoma cells can provide insights into the pathogenesis of various liver diseases, as they often display functional and metabolic alterations that mimic those observed in diseased liver tissue. These cell lines can be used to study the impact of genetic, environmental, or pharmacological interventions on liver-specific processes, such as bile acid metabolism, xenobiotic detoxification, and protein synthesis. Furthermore, some hepatoma cells exhibit characteristics of hepatic progenitor cells, allowing researchers to explore the potential of these cells for liver regeneration and cell-based therapies.
Metabolic and Signaling Pathway Investigations
Hepatoma cells also serve as models for studying dysregulated signaling cascades, such as growth factor-mediated pathways and oncogenic signaling, that contribute to the malignant phenotype of liver cancer cells. By understanding these metabolic and signaling alterations, researchers can identify novel therapeutic targets and develop strategies to selectively target the vulnerabilities of hepatoma cells.
KIF11 Negatively Correlates With Senescence Biomarkers in HCC Tissues and Hepatoma Cells
KIF11, also known as Kinesin-5, is a necessary molecular motor protein during mitosis. KIF11 mediates centromeric separation and bipolar mitotic spindle formation, thereby promoting mitosis to support cell proliferation. KIF11 is highly expressed in various malignancies, including hepatocellular carcinoma (HCC). The role of KIF11 in modulating cellular senescence has not been reported. The TCGA database and GTEx database were re-analyzed and found that KIF11 was highly expressed in HCC tissues compared with their adjacent normal tissues (Fig. 1A). Likewise, KIF11 was highly expressed in cultured hepatoma cells (HCCLM3, Huh7, HepG2, and SNU398) compared with normal liver cells (HLSEC and THLE-3) (Fig. 1B, C). Furthermore, KIF11 is negatively correlated with p16 and p14 in cultured liver cells and hepatoma cells (Fig. 1D, E). These effects were also observed in ROS stress-induced cultured normal liver cells (THLE-3) and hepatoma cells (HepG2 and Huh7) (Fig. 1F, G).
The correlation among the KIF11, p16, and p14 protein levels was analyzed in clinical HCC tissues. A total of 83 cases of HCC tissues were stained by IHC and scored according to staining intensity. As shown in the pie chart (Fig. 1H), 2 cases were KIF11high p16low, 15 cases were KIF11high p14low, 33 cases were KIF11high p16low p14low. IHC pictures showed different expressions of KIF11, p16, and p14 in clinical HCC tissues (Fig. 1I). In total, in about 60% (50 in 83 cases) of HCC patients, the expression of KIF11 is higher, while p16 or (and) p14 is low. These results indicate that the high expression of KIF11 inhibits the cellular senescence in the liver of patients with HCC. Altogether, KIF11 negatively correlates with senescence in both clinical HCC tissues and cultured hepatoma cells.
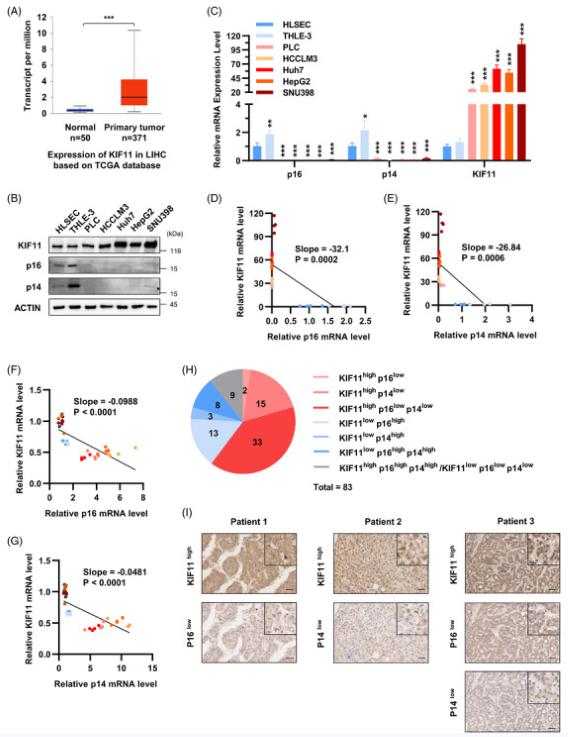 Fig.
1 Expression level of KIF11 and CDKN2A (p16 and p14) in HCC. (Chen D, et al, 2023)
Fig.
1 Expression level of KIF11 and CDKN2A (p16 and p14) in HCC. (Chen D, et al, 2023)
HRP-3 Protects the Hepatoma Cells From Glucose Deprivation-Induced Apoptosis
To estimate the long-term and stable effect of HRP-3 knockdown on HCC cells, lentivirus-based small hairpin RNA (shRNA) expression vectors (Sh1 and Sh3) were generated for two effective HRP-3-siRNA sequences (i1 and i3) and ShC as control vector for NS-siRNA. HRP-3 stable knockdown clones and control clones of SMMC-7721 (Sh1-3, Sh3-5, and ShC-7) and SK-Hep1 (Sh1-5, Sh3-10, and ShC-7), which are infected with lentivirus harboring i1, i3, and NS siRNA respectively, were constructed for further analysis. The efficiency of knockdown was detected by Western Blot (Fig. 2).
The adhesion and morphology of culture-adherent cells could give an intuitional index to cell viability and apoptosis. So the cell morphology analysis was conducted. All clones, counted for the same number and divided into two portions, were plated in 6 well plates of the same diameter with normal high-glucose DMEM overnight. One portion of each clone was changed for glucose-free DMEM the next day and the other portion was used as a control group maintained in high-glucose DMEM. After 9 hours, stable HRP-3 knockdown clones of SMMC-7721 (Sh1-5 and Sh3-10) became shrinking, round, and even suspended compared with the shC-7 control clone which had no obvious injury morphology. Similarly, results were observed between the stable HRP-3 knockdown clones of SK-Hep1 (Sh1-3 and Sh3-5) and the control clone of normal adhesion (ShC-7) after 7 hours cultured in a glucose-free medium. The morphology of all clones was captured by microscope for stochastic observation for 3 areas and the pictures were analyzed by software Image J for calculating spreading areas (Fig. 3A, B).
Then all of these clone cells were digested and submitted to flow cytometry analysis. Consistent with the morphology results, during the glucose deprivation, a progressive aggregation in the sub-G1 phase appeared in cells knocking down HRP-3 compared with in control cells (Sh1-3 and Sh3-5 Vs. ShC-7 of SMMC-7721; Sh1-5 and Sh3-10 Vs. ShC-7 of SK-Hep1), which indicates the influence of HRP-3 on cell apoptosis (Fig. 4A, B).
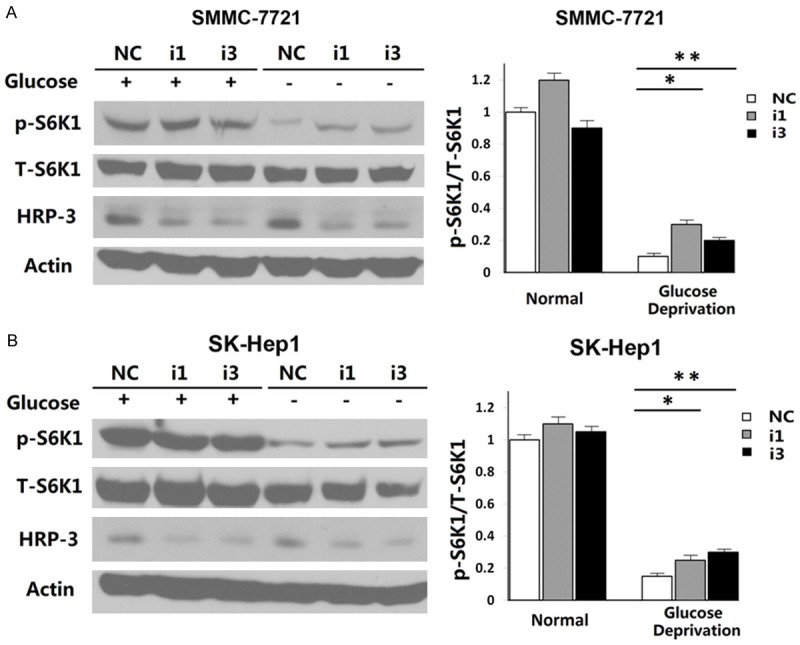 Fig.
2 Silence of HRP-3 inhibits the de-phosphorylation of S6K1 induced by glucose deprivation. (Cai H,
et al., 2015)
Fig.
2 Silence of HRP-3 inhibits the de-phosphorylation of S6K1 induced by glucose deprivation. (Cai H,
et al., 2015)
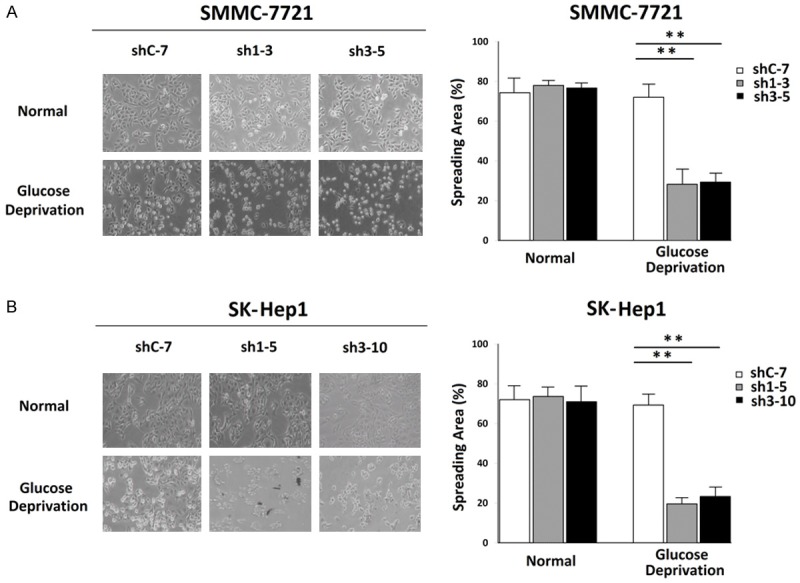 Fig.
3 Stable Knockdown of HRP-3 in HCC cells sensitized cells to become shrunk morphology of apoptosis under the
energy pressure. (Cai H, et al., 2015)
Fig.
3 Stable Knockdown of HRP-3 in HCC cells sensitized cells to become shrunk morphology of apoptosis under the
energy pressure. (Cai H, et al., 2015)
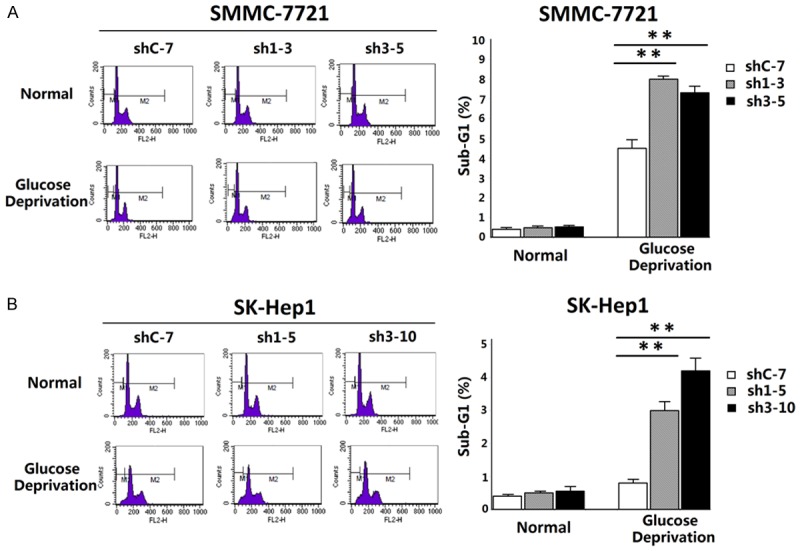 Fig.
4 HRP-3 inhibits energy deprivation-triggered apoptosis of HCC cells. (Cai H, et al., 2015)
Fig.
4 HRP-3 inhibits energy deprivation-triggered apoptosis of HCC cells. (Cai H, et al., 2015)
Hepatoma cells, particularly HepG2 cells, are commonly used in drug metabolism and hepatotoxicity studies. They retain many characteristics of normal hepatocytes (liver cells), providing a useful model for studying liver diseases and the liver's response to drugs.
Hepatoma cells typically exhibit rapid proliferation, altered metabolism, and dysregulated signaling pathways, which make them useful for studying the molecular mechanisms of liver cancer and testing potential anti-cancer therapies.
Hepatoma cells may not fully recapitulate the complex in vivo microenvironment of the liver, including interactions with other cell types and the extracellular matrix.
Filters Clear all filters
Species
- African clawed frog (1)
- American mink (1)
- Asian tiger mosquito (1)
- Atlantic salmon (1)
- Bluegill (2)
- Bluestriped grunt (1)
- Bovine (7)
- Brazilian free-tailed bat (1)
- Brown bullhead (2)
- Cabbage looper (1)
- Cabbage moth (6)
- Cat (4)
- Central mudminnow (1)
- Chicken (3)
- Chinese hamster (5)
- Chinook salmon (2)
- Chum salmon (1)
- Coho salmon (1)
- Common carp (2)
- Cotton-top tamarin (1)
- Dog (2)
- Fall armyworm (3)
- Fathead minnow (2)
- Fruit fly (1)
- Gilthead sea bream (2)
- Golden hamster (7)
- Goldfish (6)
- Gray dwarf hamster (1)
- Green monkey (2)
- Gypsy moth (1)
- Horse (1)
- Human (998)
- Japanese eel (1)
- Japanese rice fish (7)
- Koi carp (1)
- Mouse (315)
- Mouse x Gray dwarf hamster (1)
- Mouse x Rat (20)
- Northern pike (1)
- Pig (3)
- Rabbit (2)
- Rainbow trout (3)
- Rat (115)
- Rhesus macaque (1)
- Salt marsh moth (1)
- Sheep (2)
- Snakehead murrel (2)
- Sockeye salmon (1)
- Vervet monkey (2)
- Zebrafish (2)
Source
- Abdomen (1)
- Abdomen Metastasis (2)
- Adipose (2)
- Adrenal Gland (8)
- Adrenal Gland Metastasis (2)
- Aorta (4)
- Artery (1)
- Ascites (28)
- Ascites Metastasis (37)
- Bile Duct (3)
- Bladder (25)
- Bladder Metastasis (1)
- Blastocyst (1)
- Blastula (1)
- Blood (127)
- Bone (27)
- Bone Marrow (57)
- Bone Marrow Metastasis (18)
- Bone Metastasis (6)
- Brain (55)
- Brain Metastasis (8)
- Breast (30)
- Bronchus (1)
- Caudal Peduncle (1)
- Caudal Trunk (2)
- Cecum (3)
- Cerebrospinal Fluid (1)
- Cerebrospinal Fluid Metastasis (1)
- Cervix (32)
- Colon (90)
- Connective Tissue (7)
- Cornea (3)
- Cutaneous Metastasis (1)
- Dermis (2)
- Duodenum (1)
- Embryo (29)
- Endometrium (17)
- Esophagus (44)
- Eye (12)
- Eye Socket (5)
- Fetus (3)
- Fin (9)
- Foreskin (4)
- Gallbladder (1)
- Gingiva (2)
- Globe (2)
- Glomerulus (2)
- Groin (1)
- Head Kidney (2)
- Heart (4)
- Hemolymph (1)
- Hypodermis Metastasis (5)
- Ileum (1)
- Intestine (94)
- Jejunum (1)
- kidney (1)
- Kidney (27)
- Liver (35)
- Liver Metastasis (17)
- Lung (58)
- Lung Metastasis (8)
- Lymph Node (7)
- Lymph Node Metastasis (59)
- Muscle (7)
- Muscle Metastasis (2)
- Nose (2)
- Omentum Metastasis (2)
- Oral Cavity (10)
- Ovary (21)
- Ovary Metastasis (2)
- Pancreas (19)
- Pelvic Wall Metastasis (1)
- Pelvis (1)
- Perianal Space Metastasis (1)
- Pericardial Effusion (1)
- Pericardial Effusion Metastasis (2)
- Perineus (1)
- Peripheral Blood (126)
- Peripheral Nervous System (21)
- Peritoneal Effusion (2)
- Peritoneum (1)
- Peritoneum Metastasis (1)
- Pharynx (3)
- Pituitary Gland (7)
- Pleural Effusion (54)
- Pleural Effusion Metastasis (46)
- Prostate (7)
- Rectum (15)
- Renal Pelvis (1)
- Retroperitoneal Space (2)
- Salivary Gland (2)
- Skeletal Muscle (5)
- Skin (32)
- Skin Metastasis (3)
- Small Intestine (4)
- Small Intestine Metastasis (1)
- Smooth Muscle (2)
- Soft Tissue (1)
- Soft Tissue Metastasis (1)
- Spinal Cord (2)
- Stomach (4)
- Testis (15)
- Thoracic Cavity Metastasis (6)
- Thymus (5)
- Thyroid Gland (16)
- Thyroid Gland Metastasis (1)
- Tongue (5)
- Trachea (1)
- Umbilical Cord (1)
- Umbilical Cord Blood (1)
- Urachus (1)
- Ureter (1)
- Uterus (54)
- Uvea (2)
- Vagina (2)
- Vulva (1)
Disease
- Acute Biphenotypic Leukemia (1)
- Acute Erythroid Leukemia (4)
- Acute Megakaryoblastic Leukemia (4)
- Acute Monocytic Leukemia (9)
- Acute Myeloid Leukemia (25)
- Acute Promyelocytic Leukemia (2)
- Adrenal Gland Neuroblastoma (11)
- Adult B Acute Lymphoblastic leukemia (1)
- Adult B Acute Lymphoblastic Leukemia (6)
- Adult T Acute Lymphoblastic Leukemia (6)
- Adult T Lymphoblastic Lymphoma (2)
- Adult T-Cell Leukemia/Lymphoma (1)
- Alveolar Rhabdomyosarcoma (4)
- Alveolar Ridge Squamous Cell Carcinoma (1)
- Amelanotic Melanoma (3)
- Ampulla of Vater Adenocarcinoma (1)
- Ampulla of Vater Adenosquamous Carcinoma (3)
- Anaplastic Astrocytoma (3)
- Anaplastic Large Cell Lymphoma (7)
- Askin Tumor (1)
- Astrocytoma (5)
- B Acute Lymphoblastic Leukemia (2)
- B-Cell Non-Hodgkin Lymphoma (5)
- Bare Lymphocyte Syndrome Type 2 (1)
- Barrett Adenocarcinoma (2)
- Benign Prostatic Hyperplasia (1)
- Bladder Carcinoma (14)
- Bladder Squamous Cell Carcinoma (1)
- Bovine Leukemia (2)
- Breast Adenocarcinoma (4)
- Breast Carcinoma (9)
- Breast Ductal Carcinoma (2)
- Burkitt Lymphoma (17)
- Canavan Disease (1)
- Canine Histiocytic Sarcoma (1)
- Cecum Adenocarcinoma (3)
- Central Nervous System Lymphoma (2)
- Cervical Adenocarcinoma (2)
- Cervical Adenosquamous Carcinoma (2)
- Cervical Small Cell Carcinoma (1)
- Cervical Squamous Cell Carcinoma (2)
- Chicken Bursal Lymphoma (2)
- Childhood B Acute Lymphoblastic Leukemia (13)
- Childhood T Acute Lymphoblastic Leukemia (16)
- Childhood T Lymphoblastic Lymphoma (1)
- Cholangiocarcinoma (2)
- Chronic Eosinophilic Leukemia (1)
- Chronic Lymphocytic Leukemia (2)
- Chronic Myeloid Leukemia (23)
- Clear Cell Renal Cell Carcinoma (2)
- Colon Adenocarcinoma (55)
- Colon Carcinoma (34)
- Colorectal Adenocarcinoma (1)
- Colorectal Carcinoma (1)
- Congenital Pure Red Cell Aplasia (1)
- Cutaneous Melanoma (10)
- Dedifferentiated Chondrosarcoma (1)
- Desmoplastic Melanoma (1)
- Diffuse Large B-Cell Lymphoma (28)
- Down Syndrome (2)
- EBV-Related Burkitt Lymphoma (12)
- Embryonal Carcinoma (3)
- Embryonal Rhabdomyosarcoma (3)
- Endometrial Adenocarcinoma (13)
- Endometrial Adenosquamous Carcinoma (2)
- Endometrial Carcinoma (2)
- Endometrioid Stromal Sarcoma (1)
- Epithelioid Hemangioendothelioma (1)
- Epithelioid Sarcoma (3)
- Esophageal Adenocarcinoma (6)
- Esophageal Squamous Cell Carcinoma (41)
- Essential Thrombocythemia (1)
- Ewing Sarcoma (2)
- Extraskeletal Myxoid Chondrosarcoma (1)
- Fanconi Anemia (1)
- Fibrosarcoma (1)
- Follicular Lymphoma (2)
- Gallbladder Carcinoma (2)
- Gallbladder Undifferentiated Carcinoma (2)
- Gastric Adenocarcinoma (6)
- Gastric Adenosquamous Carcinoma (1)
- Gastric Carcinoma (5)
- Gastric Choriocarcinoma (1)
- Gastric Fundus Carcinoma (1)
- Gastric Signet Ring Cell Adenocarcinoma (1)
- Gastric Small Cell Carcinoma (2)
- Gastric Tubular Adenocarcinoma (5)
- Gastroesophageal Junction Adenocarcinoma (1)
- Gestational Choriocarcinoma (1)
- Gingival Squamous Cell Carcinoma (2)
- Glioblastoma (18)
- Gliosarcoma (1)
- Goldfish Erythrophoroma (4)
- Hairy Cell Leukemia (1)
- Hamster Kidney Tumor (1)
- Hamster Pancreatic Ductal Adenocarcinoma (1)
- Hamster Uterine Leiomyosarcoma (1)
- Hepatoblastoma (2)
- Hepatocellular Carcinoma (6)
- Hepatosplenic T-Cell Lymphoma (2)
- Hereditary Thyroid Gland Medullary Carcinoma (1)
- High Grade B-Cell Lymphoma (1)
- High Grade Ovarian Serous Adenocarcinoma (8)
- Hodgkin Lymphoma (9)
- Hypopharyngeal Squamous Cell Carcinoma (2)
- Infectious Mononucleosis (1)
- Intrahepatic Cholangiocarcinoma (6)
- Invasive Breast Carcinoma of No Special Type (12)
- Invasive Breast Lobular Carcinoma (1)
- Kidney Neoplasm (1)
- Kidney Rhabdoid Tumor (1)
- Krukenberg Tumor (1)
- Liposarcoma (1)
- Lung Adenocarcinoma (17)
- Lung Giant Cell Carcinoma (8)
- Lung Large Cell Carcinoma (9)
- Lung Mucoepidermoid Carcinoma (1)
- Lung Non-Small Cell Carcinoma (2)
- Lung Small Cell Carcinoma (25)
- Lung Squamous Cell Carcinoma (9)
- Lymphoblastic Lymphoma (1)
- Malignant Peripheral Nerve Sheath Tumor (1)
- Mantle Cell Lymphoma (5)
- Mature Gastric Teratoma (1)
- Maxillary Sinus Squamous Cell Carcinoma (1)
- Medaka Hepatoma (2)
- Medulloblastoma (3)
- Melanoma (24)
- Meningioma (2)
- Minimally Invasive Lung Adenocarcinoma (1)
- Monophasic Synovial Sarcoma (1)
- Mouse Bladder Transitional Cell Carcinoma (1)
- Mouse Chondrosarcoma (1)
- Mouse Colon Adenocarcinoma (3)
- Mouse Ependymoma (2)
- Mouse Erythroid Leukemia (13)
- Mouse Fibrosarcoma (5)
- Mouse Glioblastoma (1)
- Mouse Hemangioendothelioma (1)
- Mouse Hepatocellular Carcinoma (1)
- Mouse Insulinoma (3)
- Mouse Intestinal Tract Neuroendocrine Adenoma (1)
- Mouse Islet Cell Adenoma (1)
- Mouse Kidney Carcinoma (1)
- Mouse Leukemia (10)
- Mouse Leydig Cell Tumor (1)
- Mouse Lymphoma (8)
- Mouse Mammary Gland Malignant Neoplasm (23)
- Mouse Melanoma (9)
- Mouse Multiple Myeloma (5)
- Mouse Myeloid Leukemia (3)
- Mouse Neoplasm (1)
- Mouse Neuroblastoma (21)
- Mouse Oral Cavity Squamous Cell Carcinoma (1)
- Mouse Osteosarcoma (3)
- Mouse Pituitary Gland Neoplasm (1)
- Mouse Plasmacytoma (1)
- Mouse Precursor T Cell Lymphoblastic Lymphoma/Leukemia (2)
- Mouse Pulmonary Adenoma (1)
- Mouse Pulmonary Malignant Tumor (3)
- Mouse Pulmonary Squamous Cell Carcinoma (1)
- Mouse Rectum Carcinoma (2)
- Mouse Reticulum Cell Sarcoma (2)
- Mouse Sarcoma (1)
- Mouse Teratocarcinoma (8)
- Mouse Thymic Lymphoma (3)
- Mycosis Fungoides (1)
- Myelodysplastic Syndrome (1)
- Myxofibrosarcoma (1)
- Natural Killer Cell Lymphoblastic Leukemia/Lymphoma (2)
- Neuroblastoma (26)
- Oral Cavity Squamous Cell Carcinoma (15)
- Osteoid Osteoma (1)
- Osteosarcoma (15)
- Ovarian Carcinoma (1)
- Ovarian Clear Cell Adenocarcinoma (1)
- Ovarian Endometrioid Adenocarcinoma (4)
- Ovarian Granulosa Cell Tumor (1)
- Ovarian Mucinous Adenocarcinoma (2)
- Ovarian Serous Adenocarcinoma (2)
- Ovarian Serous Cystadenocarcinoma (2)
- Ovarian Small Cell Carcinoma (1)
- Pancreatic Adenocarcinoma (13)
- Pancreatic Carcinoma (5)
- Pancreatic Ductal Adenocarcinoma (12)
- Papillomavirus-Independent Cervical Squamous Cell Carcinoma (1)
- Papillomavirus-Related Cervical Adenocarcinoma (7)
- Papillomavirus-Related Cervical Squamous Cell Carcinoma (4)
- Papillomavirus-Related Endocervical Adenocarcinoma (16)
- Paroxysmal Nocturnal Hemoglobinuria (3)
- Pharyngeal Squamous Cell Carcinoma (1)
- Plasma Cell Myeloma (15)
- Pleural Epithelioid Mesothelioma (5)
- Pleural Sarcomatoid Mesothelioma (2)
- Poorly Differentiated Thyroid Gland Carcinoma (1)
- Primary Cutaneous T-Cell Non-Hodgkin Lymphoma (1)
- Primary Effusion Lymphoma (7)
- Primitive Neuroectodermal Tumor (1)
- Prostate carcinoma (1)
- Prostate Carcinoma (9)
- Rat C-Cell Carcinoma (1)
- Rat Cholangiocarcinoma (1)
- Rat Colon Adenocarcinoma (5)
- Rat Digestive System Neoplasm (1)
- Rat Fibrosarcoma (1)
- Rat Hepatocellular Carcinoma (20)
- Rat Histiocytic Sarcoma (1)
- Rat Insulinoma (2)
- Rat Leukemia (1)
- Rat Leydig Cell Adenoma (1)
- Rat Lung Carcinoma (1)
- Rat Malignant Glioma (4)
- Rat Malignant Meningioma (1)
- Rat Malignant Oligodendroglioma (2)
- Rat Malignant Thymoma (3)
- Rat Mammary Gland Adenocarcinoma (10)
- Rat Neuroblastoma (3)
- Rat Osteosarcoma (2)
- Rat Pituitary Gland Neoplasm (6)
- Rat Prostate Adenocarcinoma (3)
- Rat Rhabdomyosarcoma (1)
- Rat Sarcoma (2)
- Rat Squamous Cell Carcinoma (1)
- Rat Urinary Bladder Transitional Cell Carcinoma (2)
- Rat Urinary System Neoplasm (6)
- Rectal Adenocarcinoma (13)
- Rectosigmoid Adenocarcinoma (1)
- Recurrent Bladder Carcinoma (1)
- Renal Cell Carcinoma (7)
- Renal Pelvis Urothelial Carcinoma (1)
- Retinoblastoma (11)
- Sacral Chordoma (1)
- Sacrococcygeal Teratoma (1)
- Salivary Gland Squamous Cell Carcinoma (1)
- Sezary Syndrome (1)
- Shwachman-Diamond Syndrome (1)
- Skin Squamous Cell Carcinoma (2)
- Splenic Marginal Zone Lymphoma (1)
- Testicular Embryonal Carcinoma (8)
- Testicular Teratoma (2)
- Testicular Yolk Sac Tumor (1)
- Thyroid Gland Anaplastic Carcinoma (10)
- Thyroid Gland Follicular Carcinoma (4)
- Thyroid Gland Papillary Carcinoma (3)
- Thyroid Gland Sarcoma (1)
- Thyroid Gland Squamous Cell Carcinoma (2)
- Tongue Adenosquamous Carcinoma (1)
- Tongue Squamous Cell Carcinoma (6)
- Type I Endometrial Adenocarcinoma (1)
- Ureter Urothelial Carcinoma (1)
- Uterine Carcinosarcoma (2)
- Uterine Corpus Leiomyosarcoma (1)
- Uterine Corpus Sarcoma (2)
- Uveal Melanoma (2)
- Vaginal Melanoma (2)
- Vulvar Melanoma (1)
- Vulvar Squamous Cell Carcinoma (1)
Description: Established from the tumor tissue of an 8-year-old black boy in 1976.The cells contain a 2.3 kb ...
Description: The Doxorubicin-resistant cell line HepG2/ADR has been developed by repeatedly exposing the parent ...
Description: Human cell line derived from cholangiocellular carcinoma.
Description: A subline of HepG2. Expressing abundant glutamine synthetase.
Description: Human cell line derived from cholangiocellular carcinoma. Cell growth is slow.
Description: Human bile duct cell line established from ascites of the tumor patient who had differentiated ...
Description: Ito (fat-storing) cells, stellate-shaped mesenchymal cells that exist in the space of Disse of the ...
Description: Human hepatocellular carcinoma (Ed.I) established from small liver cancer.
Description: This is one cell line out of a series of glioblastoma cell lines established by PD Dr. Michael ...
Description: This line contains an integrated hepatitis B virus genome.
Description: Human liver bile duct carcinoma cell line.