Telomere Length Analysis (Q-FISH)
- Service Details
- Features
- FAQ
- Explore Other Options
Telomeres are repetitive, non-coding sequence segments located at the termini of eukaryotic chromosomes, playing a crucial role in safeguarding chromosomal structural integrity. Due to the end-replication problem encountered during cell division, telomere length progressively shortens. When telomeres reach a critical length threshold, cells initiate senescence mechanisms. Consequently, telomere length is regarded as a vital biomarker of cellular aging. Advances in research have underscored the importance of precise telomere length measurement for understanding cellular senescence and related disease mechanisms.
Currently, Q-FISH (quantitative fluorescence in situ hybridization) technology, with its high sensitivity and high resolution, has become an important method for telomere length detection. This technique utilizes fluorescently labeled PNA (peptide nucleic acid) probes to measure telomere length through quantitative analysis of fluorescent signal intensity. It can even be applied to paraffin-embedded tissue samples, enabling detection at the single-cell level.
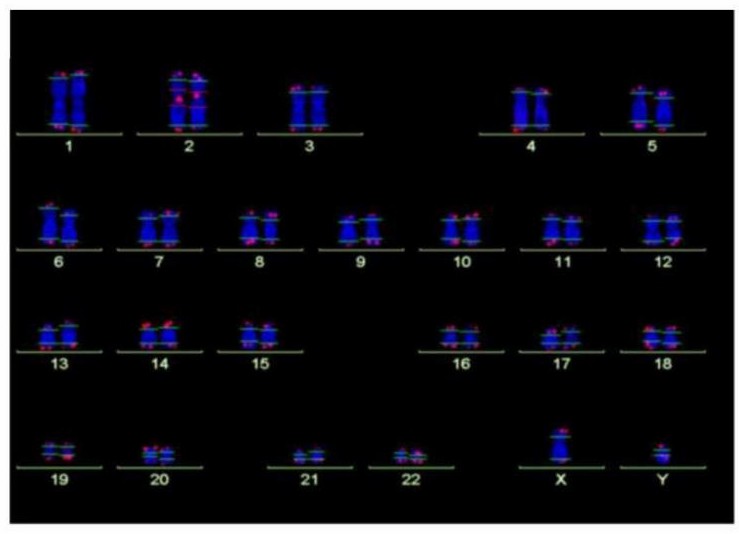 Fig. 1. Analysis of telomere length by Q-FISH. Normal karyogram with single telomeres stained for telomere repeats (Q-FISH) and a repetitive region in the centromeric region of chromosome 2 (Habib R, Kim R, et al., 2020).
Fig. 1. Analysis of telomere length by Q-FISH. Normal karyogram with single telomeres stained for telomere repeats (Q-FISH) and a repetitive region in the centromeric region of chromosome 2 (Habib R, Kim R, et al., 2020).
Advantages of Q-FISH for telomere length measurement:
- Single-cell precision: Accurately measures telomere length at the individual chromosome-arm level.
- Broad sample compatibility: Compatible with fresh cell samples and FFPE tissue sections.
- Morphological integration: Enables simultaneous visualization of cellular morphology and telomere length under microscopy.
Our Q-FISH Service
Workflow
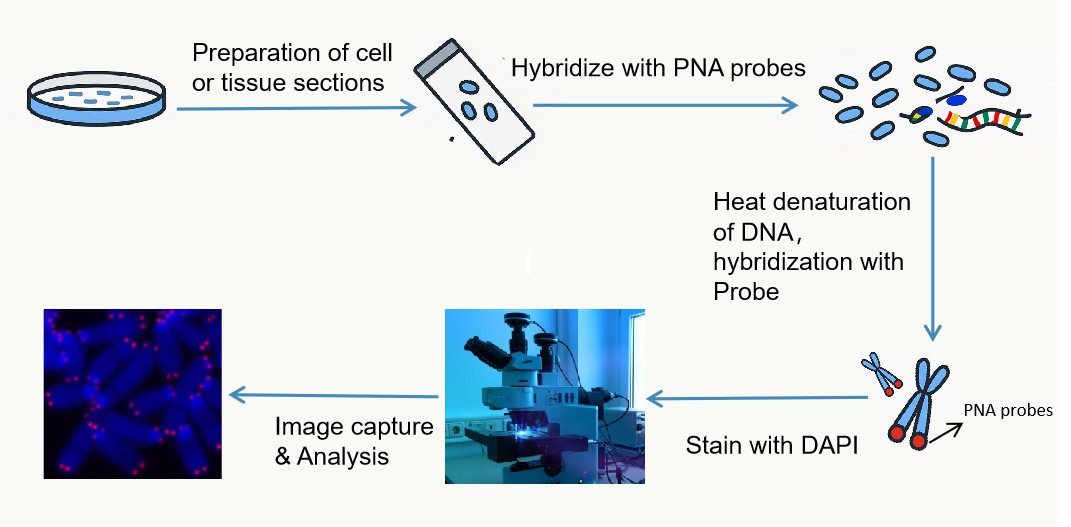 Fig. 2. Basic procedure of Q-FISH.
Fig. 2. Basic procedure of Q-FISH.
- Consultation & Needs Assessment: Collaborate with clients to define research objectives and sample specifications.
- Sample Receiving & Processing: Quality evaluation (cell viability/tissue integrity testing).
- Q-FISH Experiment: Hybridization with fluorescent PNA probes and high-resolution fluorescence imaging.
- Data Analysis: Telomere signal quantification using specialized software.
- Report Delivery: Comprehensive analytical report with interpretable results.
Sample Types
- In vitro cultured cells
- Peripheral blood mononuclear cells (PBMCs)
- Frozen or fresh tissue sections (meeting thickness requirements)
Detection Metrics
- Mean telomere length
- Short telomeres
Key Features

High precision
Advanced Q-FISH technology combined with high-resolution fluorescence microscopy and professional image analysis software.

Expert team
Staffed by experienced molecular biologists and biotechnologists.

Customized solutions
Tailored protocols for diverse research needs.

Rapid turnaround
Streamlined workflows and efficient data analysis.
FAQ
1. Why is Q-FISH superior to other techniques?
- Single-cell resolution: Identifies telomere length heterogeneity within cell populations (e.g., extreme telomere length variation in tumor subclones).
- High sensitivity: Compatible with low-abundance samples (e.g., microtissues, circulating tumor cells [CTCs]), offering a 40% lower error rate compared to qPCR.
- Multiparametric analysis: Enables correlation of telomere length with cell proliferation (e.g., Ki67) or DNA damage markers (e.g., γH2AX).
2. What sample types are acceptable? Can frozen samples be analyzed?
We accept diverse sample types, including fresh cells, frozen cells, and tissue sections. For frozen cells, pre-freezing viability must be ≥80%. Specific handling protocols are provided based on sample characteristics.
3. What are the potential sources of experimental error, and how can they be managed or mitigated?
- Major error:
Sample quality: Low cell viability or improper tissue fixation may degrade telomeric DNA.
Probe specificity: Non-specific hybridization may overestimate telomere length.
Imaging parameters: Inconsistent exposure time or fluorescence gain settings affect quantification.
- Quality control measures:
Inclusion of positive controls (immortalized cell lines with stable telomere length) and negative controls (RNase-treated samples) in each batch.
Double-blind verification to ensure ≥95% data reproducibility.
Reference
- Habib R, Kim R, et al. Telomere attrition and dysfunction: a potential trigger of the progeroid phenotype in nijmegen breakage syndrome. Aging (Albany NY). 2020. 12(12):12342-12375
Explore Other Options