Comparative Genomic Hybridization and Its Applications
Comparative genomic hybridization (CGH) is a technique that permits the detection of chromosomal copy number changes without the need for cell culturing. Simultaneous hybridization of differentially labeled test DNA and normal reference DNA with normal metaphase chromosomes can provide a global overview of chromosome gains and losses in the entire genome. It allows the analysis of the entire genome in a single experiment. CGH is based on the principles of fluorescent in situ hybridization (FISH). The result of the hybridization is imaged with a fluorescence microscope, karyotyped, and computerized.
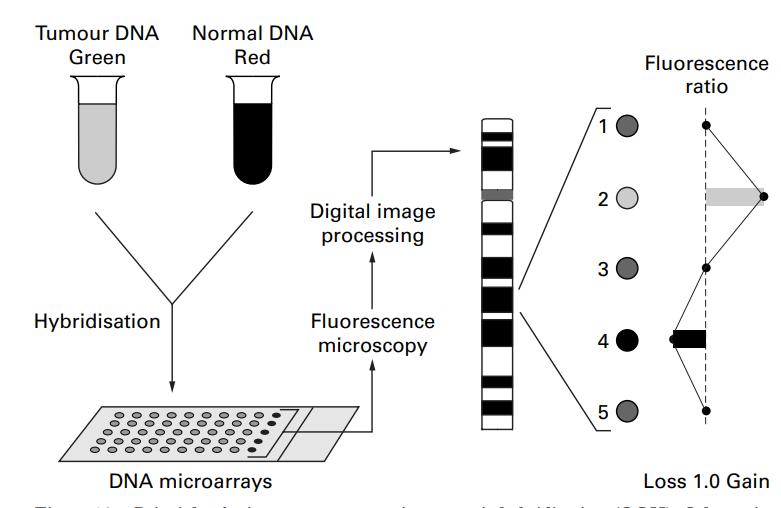 Fig.1 Principle of microarray CGH. (Weiss MM, et al., 1999)
Fig.1 Principle of microarray CGH. (Weiss MM, et al., 1999)
Traditional & Micro CGH Analysis
- Comparative genomic hybridization (CGH) can be used for comprehensive low-resolution analysis of chromosomal imbalances of an entire human genome. The so-called microdissection-based comparative genomic hybridization (micro-CGH) technique allows the molecular cytogenetic investigation of harvested and cytogenetically fixed interphase nuclei. The CGH method is mainly used in cytogenetics for the detection of unbalanced chromosomal aberration in tumors.
- Creative Bioarray provides a platform for traditional CGH analysis and optional micro-CGH analysis based on FISH services. This service can be used for cytogenetic analysis of solid tumor samples. The basic process of traditional CGH analysis includes sample preparation, probe preparation, FISH hybridization, staining, imaging, and data analysis. In addition, we provide independent micro-CGH services, and we have independent experiments to complete chromosome microdissections.
High-Resolution CGH
- It differs from classical CGH in the way of software processing of the captured fluorescence image. It uses dynamic standard reference intervals (natural variation of fluorescence intensities of chromosome pairs) instead of fixed ones. Dynamic values are obtained by statistically processing the deviations of control profiles in individuals with a normal karyotype.
- The advantage of the method is the resolution at the level of 4-5 Mb and the possibility of detecting aberrations even at 20–30% clonal representation.
Array CGH Analysis
- Hybridization of fluorescently labeled probes of the DNA under investigation and reference DNA with many specific DNA sequences (instead of chromosomes). The genome is thus divided into small sections and arranged in a grid (microarray, biochip). The individual sections have a precise location and can thus represent individual genes or parts of genes.
- Array CGH has many research applications including cancer profiling, gene discovery, and understanding epigenetic modifications and chromatin conformation. The results from such investigations can be directly correlated to genomic locations and gene expression.
- Creative Bioarray provides Array CGH Services, a high-resolution karyotype analysis solution, for the detection of unbalanced structural and numerical chromosomal alterations with high-throughput capabilities. In addition, a wide range of Karyotyping Services (Traditional Karyotyping-G Banded, M-FISH, Molecular Karyotyping, etc.) is provided to meet our customer's different needs.
Reference
- Weiss MM, et al. (1999). "Comparative genomic hybridization." Mol Pathol. 52 (5), 243-51.
For research use only. Not for any other purpose.
Resources
- FAQ
- Protocol
- Cell Culture Guide
- Technical Bulletins
-
Explore & Learn
-
Cell Biology
- How to Handle Mycoplasma in Cell Culture?
- CHO Cell Line Development
- Troubleshooting Cell Culture Contamination: A Comprehensive Guide
- How to Isolate PBMCs from Whole Blood?
- Monocytes vs. Macrophages
- How to Detect and Remove Endotoxins in Biologics?
- Comparison of Different Methods to Measure Cell Viability
- How to Isolate and Analyze Tumor-Infiltrating Leukocytes?
- Contamination of Cell Cultures & Treatment
- Generation and Applications of Neural Stem Cells
- Stem Cell Markers
- Comparison of the MSCs from Different Sources
- T Cell Activation and Expansion
- Organoid Differentiation from Induced Pluripotent Stem Cells
- Quantification of Cytokines
- Tips For Cell Cryopreservation
- What are the Differences Between M1 and M2 Macrophages?
- Mesenchymal Stem Cells: A Comprehensive Exploration
- What Cell Lines Are Commonly Used in Biopharmaceutical Production?
- How to Assess the Migratory and Invasive Capacity of Cells?
- Enrichment, Isolation and Characterization of Circulating Tumor Cells (CTCs)
- Strategies for Enrichment of Circulating Tumor Cells (CTCs)
- How to Decide Between 2D and 3D Cell Cultures?
- Isolation, Expansion, and Analysis of Natural Killer Cells
- Neural Differentiation from Induced Pluripotent Stem Cells
- Circulating Tumor Cells as Cancer Biomarkers in the Clinic
- CFU Assay for Hematopoietic Cell
- Biomarkers and Signaling Pathways in Tumor Stem Cells
- Techniques for Cell Separation
- How to Start Your Culture: Thawing Frozen Cells
- What Are Myeloid Cell Markers?
- Cryopreservation of Cells Step by Step
- What are PBMCs?
- How to Eliminate Mycoplasma From Cell Cultures?
- Cell Cryopreservation Techniques and Practices
- Guidelines for Cell Banking to Ensure the Safety of Biologics
- Critical Quality Attributes and Assays for Induced Pluripotent Stem Cells
- T Cell, NK Cell Differentiation from Induced Pluripotent Stem Cells
- What Is Cell Proliferation and How to Analyze It?
- Cell Culture Medium
- Major Problems Caused by the Use of Uncharacterized Cell Lines
- Multi-Differentiation of Peripheral Blood Mononuclear Cells
- STR Profiling—The ID Card of Cell Line
- Comparison of Several Techniques for the Detection of Apoptotic Cells
- Human Primary Cells: Definition, Assay, Applications
- What are Mesothelial Cells?
- How to Scale Up Single-Cell Clones?
- Unveiling the Molecular Secrets of Adipogenesis in MSCs
- Tumor Stem Cells: Identification, Isolation and Therapeutic Interventions
- Direct vs. Indirect Cell-Based ELISA
- IL-12 Family Cytokines and Their Immune Functions
- Spheroid vs. Organoid: Choosing the Right 3D Model for Your Research
- From Collection to Cure: How ACT Works in Cancer Immunotherapy
- Understanding Immunogenicity Assays: A Comprehensive Guide
- Role of Cell-Based Assays in Drug Discovery and Development
- How to Maximize Efficiency in Cell-Based High-Throughput Screening?
- Immunogenicity Testing: ELISA and MSD Assays
- Types of Cell Therapy for Cancer
- What are White Blood Cells?
- 3D-Cell Model in Cell-Based Assay
- Immunogenicity Testing: ELISA and MSD Assays
- What Are the Pros and Cons of Adoptive Cell Therapy?
- Eosinophils vs. Basophils vs. Neutrophils
- 3D-Cell Model in Cell-Based Assay
- Cultivated Meat: What to Know?
- Cell Viability, Proliferation and Cytotoxicity Assays
- What Are CAR T Cells?
- Exploring Cell Dynamics: Migration, Invasion, Adhesion, Angiogenesis, and EMT Assays
- From Blur to Clarity: Solving Resolution Limits in Live Cell Imaging
- A Complete Guide to Immortalized Cancer Cell Lines in Cancer Research
- Optimization Strategies of Cell-Based Assays
- From Primary to Immortalized: Navigating Key Cell Lines in Biomedical Research
- From Blur to Clarity: Solving Resolution Limits in Live Cell Imaging
- Key Techniques in Primary, Immortalized and Stable Cell Line Development
- Live Cell Imaging: Unveiling the Dynamic World of Cellular Processes
- Optimization Strategies of Cell-Based Assays
- Overview of Cell Apoptosis Assays
- Cell-Based High-Throughput Screening Techniques
- Adherent and Suspension Cell Culture
- Mastering Cell Culture and Cryopreservation: Key Strategies for Optimal Cell Viability and Stability
- Cell Immortalization Step by Step
- Live Cell Imaging: Unveiling the Dynamic World of Cellular Processes
-
Histology
- Fluorescent Nuclear Staining Dyes
- Stains Used in Histology
- Troubleshooting in Fluorescent Staining
- Guides for Live Cell Imaging Dyes
- Overview of the FFPE Cell Pellet Product Lines
- Immunohistochemistry Controls
- Instructions for Tumour Tissue Collection, Storage and Dissociation
- Tips for Choosing the Right Protease Inhibitor
- Mitochondrial Staining
- Overview of Common Tracking Labels for MSCs
- How to Apply NGS Technologies to FFPE Tissues?
- Multiple Animal Tissue Arrays
- Immunohistochemistry Troubleshooting
- Cell and Tissue Fixation
- Cell Lysates: Composition, Properties, and Preparation
- Comparison of Membrane Stains vs. Cell Surface Stains
- Microscope Platforms
- How to Choose the Right Antibody for Immunohistochemistry (IHC)
- How to Begin with Multiplex Immunohistochemistry (mIHC)
- Common Immunohistochemistry Stains and Their Role in Cancer Diagnosis
- Serum vs. Plasma
- Comparing IHC, ICC, and IF: Which One Fits Your Research?
- What You Must Know About Neuroscience IHC?
- Modern Histological Techniques
- Multiplexing Immunohistochemistry
- How Immunohistochemistry Makes the Invisible Brain Visible?
- Histological Staining Techniques: From Traditional Chemical Staining to Immunohistochemistry
- From Specimen to Slide: Core Methods in Histological Practice
-
Exosome
- What's the Potential of PELN in Disease Treatment?
- Classification, Isolation Techniques and Characterization of Exosomes
- Emerging Technologies and Methodologies for Exosome Research
- How to Label Exosomes?
- How to characterize exosomes?
- How to Enhancement Exosome Production?
- How to Efficiently Utilize MSC Exosomes for Disease Treatment?
- How to Perform Targeted Modification of Exosomes?
- Exosomes as Emerging Biomarker Tools for Diseases
- How to Apply Exosomes in Clinical?
- Exosome Transfection for Altering Biomolecular Delivery
- Summary of Approaches for Loading Cargo into Exosomes
- Exosome Antibodies
- Collection of Exosome Samples and Precautions
- Current Research Status of Milk Exosomes
- How do PELN Deliver Drugs?
- Techniques for Exosome Quantification
- How Important are Lipids in Exosome Composition and Biogenesis?
- Common Techniques for Exosome Nucleic Acid Extraction
- What are the Functions of Exosomal Proteins?
- Exosome Size Measurement
- The Role of Exosomes in Cancer
- Exosome Quality Control: How to Do It?
- Unraveling Biogenesis and Composition of Exosomes
- Applications of MSC-EVs in Immune Regulation and Regeneration
- Production of Exosomes: Human Cell Lines and Cultivation Modes
-
ISH/FISH
- ISH probe labeling method
- Reagents Used in FISH Experiments
- Multiple Approaches to Karyotyping
- Comprehensive Comparison of IHC, CISH, and FISH Techniques
- RNAscope ISH Technology
- CARD-FISH: Illuminating Microbial Diversity
- What are the Differences between FISH, aCGH, and NGS?
- What are Single, Dual, and Multiplex ISH?
- Comparative Genomic Hybridization and Its Applications
- Overview of Oligo-FISH Technology
- Differences Between DNA and RNA Probes
- FISH Tips and Troubleshooting
- What Is the Use of FISH in Solid Tumors?
- Mapping of Transgenes by FISH
- Multiple Options for Proving Monoclonality
- FISH Techniques for Biofilm Detection
- Whole Chromosome Painting Probes for FISH
- Telomere Length Measurement Methods
- In Situ Hybridization Probes
- Overview of Common FISH Techniques
- Guidelines for the Design of FISH Probes
- Different Types of FISH Probes for Oncology Research
- Small RNA Detection by ISH Methods
- What Types of Multicolor FISH Probe Sets Are Available?
- How to Use FISH in Hematologic Neoplasms?
- ImmunoFISH: Integrates FISH and IL for Dual Detection
- 9 ISH Tips You Can't Ignore
-
Toxicokinetics & Pharmacokinetics
- Pharmacokinetics of Therapeutic Peptides
- What Are Metabolism-Mediated Drug-Drug Interactions?
- How to Improve Drug Plasma Stability?
- How Is the Cytotoxicity of Drugs Determined?
- Toxicokinetics vs. Pharmacokinetics
- How to Improve the Pharmacokinetic Properties of Peptides?
- Pharmacokinetics Considerations for Antibody Drug Conjugates
- Key Considerations in Toxicokinetic
- Organoids in Drug Discovery: Revolutionizing Therapeutic Research
- Experimental Methods for Identifying Drug-Drug Interactions
- How to Conduct a Bioavailability Assessment?
- Organ-on-a-Chip Systems for Drug Screening
- Traditional vs. Novel Drug Delivery Methods
- What factors influence drug distribution?
- How to Design and Synthesize Antibody Drug Conjugates?
- What Is the Role of the Blood-Brain Barrier in Drug Delivery?
- Parameters of Pharmacokinetics: Absorption, Distribution, Metabolism, Excretion
- What are the Pharmacokinetic Properties of the Antisense Oligonucleotides?
- Methods of Parallel Artificial Membrane Permeability Assays
- Key Factors Influencing Brain Distribution of Drugs
- Comparison of MDCK-MDR1 and Caco-2 Cell-Based Permeability Assays
- Unraveling the Role of hERG Channels in Drug Safety
- The Rise of In Vitro Testing in Drug Development
- Predictive Modeling of Metabolic Drug Toxicity
- Overview of In Vitro Permeability Assays
- Physical and Chemical Properties of Drugs and Calculations
- How to Improve Drug Distribution in the Brain
- What Are Compartment Models in Pharmacokinetics?
- Effects of Cytochrome P450 Metabolism on Drug Interactions
- When Should You Introduce ADME Tox Testing in Drug Development?
- From Cells to Systems: Modern Approaches to Disease Modeling
- How to Choose the Right In Vitro ADME Assays for Small-Molecule Drugs
- How Genotoxicity Testing Guides Safer Drug Development
- Top 5 Pitfalls in In Vitro ADME Assays and How to Avoid Them
- What Is Genotoxicity in Pharmacology? Mechanisms and Sources
- In Vitro ADME vs In Vivo ADME
- Why Cardiotoxicity Matters in R&D?
- What Are the Best Methods to Test Cardiotoxicity?
-
Disease Models
- Preclinical Models of Acute Liver Failure
- What Human Disease Models Are Available for Drug Development?
- Overview of Cardiovascular Disease Models in Drug Discovery
- Animal Models of Neurodegenerative Diseases
- Summary of Advantages and Limitations of Different Oncology Animal Models
- Disease Models of Diabetes Mellitus
- Why Use PDX Models for Cancer Research?
-
Cell Biology
- Life Science Articles
- Download Center
- Trending Newsletter