Bacterial Reverse Mutation Test (Ames Test, OECD 471)
- Service Details
- Features
- Applications
- FAQ
The Ames Test is a cornerstone method for genotoxicity screening, efficiently predicting the potential carcinogenic risk of chemicals by detecting their ability to induce genetic mutations in specific bacterial strains. In drug development, chemical safety, and environmental risk assessment, the Ames Test is highly valued for its sensitivity, speed, and cost-effectiveness, making it a mandatory core test required by global regulatory agencies such as the FDA, EMA, and EPA. Early identification of mutagenicity significantly reduces the risks of later-stage development, helping to prevent clinical trial failures or product recalls due to genotoxic issues.
Creative Bioarray leverages years of experience and technological innovation to offer you bacterial reverse mutation testing services in compliance with international standards. Our evaluation process adheres to the OECD 471 guideline by using engineered Salmonella typhimurium and Escherichia coli strains in combination with the metabolic activation system (S9 Mix) to create mammalian metabolic conditions which ensures both scientific accuracy and evaluation reliability.
Creative Bioarray's Ames Test:
Workflow
1
Requirement Confirmation and Solution Design
Tailor experimental plans based on client needs (e.g., test substance type, concentration range).
2
Strain Selection and Preparation
Use multiple strains of Salmonella typhimurium and Escherichia coli covering various mutation types like base-pair substitution and frameshift mutations.
3
Sample Processing
Mix the test compound with bacterial strains, incorporating a metabolic activation system (such as S9 mix) to simulate in vivo metabolism.
4
Culturing and Observation
Co-culture test substances with strains on histidine/tryptophan-deficient agar plates and observe the number of revertant colonies.
5
Data Collection and Analysis
Count the number of revertant colonies and assess the mutagenic potential of the test substance via dose-response relationships.
6
Report Delivery
Provide a detailed GLP-compliant report, including experimental methods, result interpretation, and conclusion recommendations.
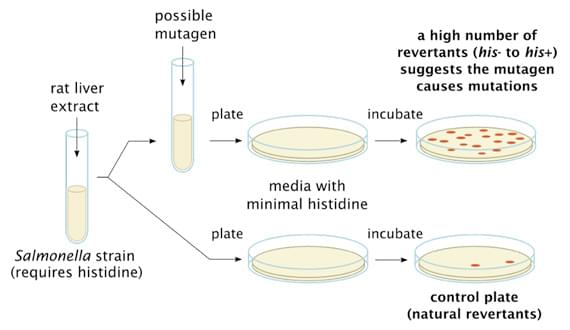 Fig. 1. General Procedure of Ames Test.
Fig. 1. General Procedure of Ames Test.
| Ames Test | Details |
| Method |
Ames Test, OECD 471
|
| Strains |
|
| Test Article Concentrations |
Eight different dose levels available Maximum concentration: 5 mg/plate (or maximum soluble concentration) |
| Metabolizing System |
|
| Controls |
|
| Delivery | Report including experimental methods, raw colony counts, dose-response curves, statistical analysis (t-test), conclusion determination |
We also offer the Mini-Ames assay, a development from the standard Ames test, which significantly reduces the quantity of test substance required (300 mg) to accelerate the drug discovery process.
Detection Scope:
Test Substance Types:
- chemical raw materials;
- drug candidate molecules;
- cosmetic ingredients;
- industrial chemicals;
- environmental pollutants, etc.
Mutation Types Detected:
- base substitution mutations;
- frameshift mutations;
- mutations related to oxidative damage.
Advantages of Our Ames Test Services

International Compliance
Strict adherence to OECD 471 international standards; data usable for global regulatory submissions.

Flexible Plan Design
Support for tailored testing conditions (e.g., managing high-concentration insoluble substances, meeting unique metabolic activation needs).

Rapid Delivery Cycle
Fast testing cycle, from sample submission to result delivery in just a few days.

Comprehensive Reporting
Detailed experimental reports including analytical methods, raw data, and result interpretations.
Applications
- Drug Development: Early screening of candidate compound genotoxicity to reduce clinical trial risk.
- Chemical Registration: Required testing to comply with REACH and pesticide registration regulations.
- Environmental Safety Assessment: Detect potential mutagenicity of industrial wastewater and soil pollutants.
- Consumer Product Safety: Evaluate the genetic toxicity risks of cosmetics, food additives, and other daily products.
Contact Us
For more information or to request a quote, get in touch with us. Our toxicology experts will provide personalized technical consultation!
FAQ
What makes studying compound mutagenicity crucial?
Studying the mutagenicity of compounds is extremely important as it directly impacts public health and safety. When a compound exhibits mutagenicity it damages DNA which may result in cancer or genetic disorders. Global regulatory agencies including the FDA and EPA require mutagenicity testing as a mandatory procedure for new drugs and chemicals before they reach consumers. Testing for mutagenicity helps organizations fulfill regulatory standards and prevents expensive market withdrawals or legal claims caused by unrecognized genetic toxicity that surfaces during later development stages.
What is the difference between the plate incorporation and pre-incubation methods in the Ames Test?
The plate incorporation method requires test substances to be combined directly with agar while the pre-incubation method needs bacteria to interact with the test substance during a fixed incubation period before being mixed. The pre-incubation method works best for unstable or volatile compounds because it enables more effective interactions between the compound and bacteria in liquid conditions which improves sensitivity detection.
How is a positive result determined?
The main criterion for a positive outcome in the Ames test involves observing a substantial growth in revertant colonies. The test sample treatment group should show a minimum of twice as many revertant colonies compared to the control group while demonstrating a dose-response relationship when the dose increases. Appropriate statistical analysis such as a t-test is necessary to verify this statistical significance.
Why does mutagenicity testing in the Ames Test require at least five test strains?
Using at least five test strains in the Ames Test ensures comprehensive coverage of different types of DNA damage and repair pathways. This includes strains sensitive to base-pair substitutions, frameshift mutations, and more. The use of multiple strains increases the test's breadth and reduces the risk of false negatives, as different strains exhibit different sensitivities to various mutagenic mechanisms.
Explore Other Options