Telomere Length Measurement Methods
Telomeres are highly conserved tandem nucleotide repeats that include proximal double-stranded and distal single-stranded regions that in complex with shelterin proteins afford protection at chromosomal ends to maintain genomic integrity. It is also generally accepted that progressive telomere shortening can lead to in vitro replicative senescence and, in the presence of other alterations, short telomeres can form bridge-fusion-breakage cycles leading to chromosome instability and the development of cancer. Here, we describe several major methods developed to measure telomere lengths (TLs) in cells and tissues.
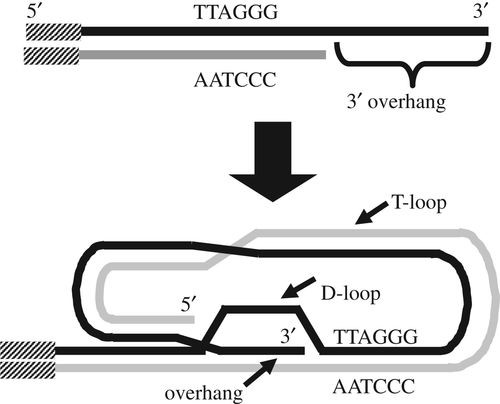 Fig.1 A model of a typical telomere. (Lai TP, et al., 2018)
Fig.1 A model of a typical telomere. (Lai TP, et al., 2018)
Terminal Restriction Fragmentation
- Based on the knowledge that the sequence TTAGGGn was highly conserved, the TRF analysis was developed using a TTAGGGn-labelled probe and is now a widely used method for telomere measurement. Regarded as the "gold standard" for TL measurement, TRF analysis measures the intensity of telomere smears to generally determine an average TL.
- In this procedure, genomic DNA is exhaustively digested using a cocktail of frequent cutting restriction enzymes that lack recognition sites in the telomeric and subtelomeric regions (and hence do not "cut" telomeric DNA). The intact telomeres from all chromosomes are then resolved, based on size, using agarose gel electrophoresis, with the telomeric fragments being visualized by either southern blotting or in-gel hybridization using a probe specific for telomeric DNA.
Polymerase Chain Reaction-based Techniques (qPCR, MMqPCR, and aTL qPCR)
- To overcome the hurdle of needing large quantities of DNA to evaluate telomere lengths, polymerase chain reaction (PCR)-based telomere length analysis methods have been developed. These procedures include quantitative (or real-time) PCR (qPCR), monochrome multiplex quantitative PCR (MMqPCR), and absolute telomere length (aTL) quantitation.
- Typically, in qPCR, the amount of the DNA sequence of interest is quantified through the use of a fluorophore (which emits a fluorescent signal) that intercalates with double-stranded DNA (i.e., SYBR green), or a probe with an attached fluorophore that is released when the sequence of interest is amplified (i.e., TaqMan probes). After each cycle, the amount of emitted fluorescence is measured, allowing the quantity of starting material to be inferred.
Quantitative Fluorescence in Situ Hybridization
- Quantitative fluorescence in situ hybridization (Q-FISH) of telomeric repeats is performed by assessing metaphase chromosomes or interphase nuclei following hybridization/labeling with a fluorescent (CCCTAA)3 probe. Unlike the TRF and PCR-based assays, the substrate for Q-FISH is cells (rather than DNA).
- Creative Bioarray provides Q-FISH-based telomere analysis, to help our customers study molecular mechanisms related to telomere length regulation, cell cycle, cell lifespan, and more. Apart from that, we also offer Metaphase FISH Service, Interphase FISH Service, and Flow-FISH Service. These methods are based on similar principles but with some modifications for different applications.
References
- Lai TP, et al. (2018). "Comparison of telomere length measurement methods." Philos Trans R Soc Lond B Biol Sci. 373 (1741), 20160451.
- Lai TP, Wright WE, Shay JW. Comparison of telomere length measurement methods. Philos Trans R Soc Lond B Biol Sci. 2018 Mar 5;373(1741):20160451.