Spatial Metabolomics Solutions
- Service Details
- Workflow
- Features
- Applications
- FAQ
Deciphering the Spatial Metabolic Code of Life
Metabolomics, a cornerstone of systems biology, serves as the most direct window into phenotypic dynamics. The synthesis and accumulation of metabolites exhibit strict spatial specificity, with their physiological functions intimately linked to their localization within tissues, cells, and even subcellular compartments. Traditional metabolomics, limited by homogenized sample processing, inherently loses critical spatial information. In contrast, spatial metabolomics bridges this gap by integrating mass spectrometry imaging (MSI) technology with multidimensional metabolic analysis, extending research dimensions to two- or three-dimensional spatial distributions. This advancement enables precise unraveling of the in situ regulatory mechanisms governing metabolic networks in biological tissues.
Our Spatial Metabolomics Service
Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry Imaging (MALDI-MSI) stands as the gold-standard technology for spatial metabolomics research. By utilizing laser excitation to activate the matrix-sample co-crystalline layer, this technique enables in situ desorption/ionization of metabolites. Coupled with high-precision mass spectrometry scanning and intelligent imaging algorithms, MALDI-MSI simultaneously captures molecular structures, abundances, and spatial distribution information of hundreds of metabolites at micrometer-level resolution. Compared to conventional imaging technologies, MALDI-MSI offers three transformative advantages:
- Label-Free Analysis: No labels required, detects over 2,000 metabolites like lipids and drugs in a single run.
- Unmatched Precision: Advanced ion sources and laser systems achieve 5 μm resolution, revealing metabolic differences at single-cell levels.
- Multimodal Compatibility: Integrates with H&E staining and IHC images, and allows combined metabolomic and proteomic analysis on the same sample.
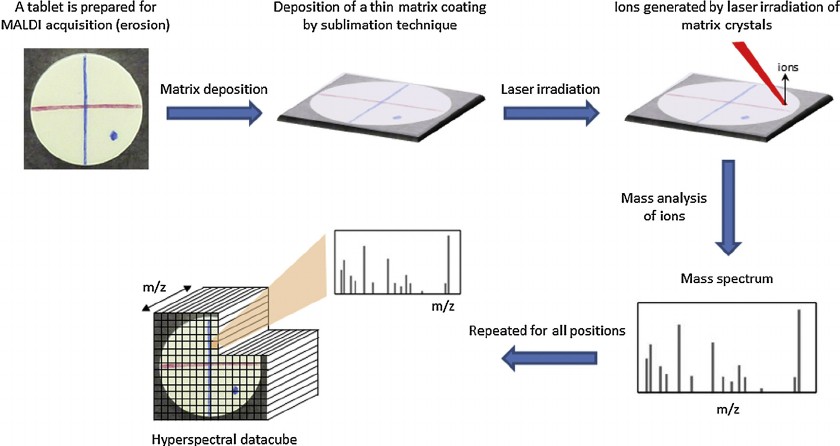 Fig. 1. Representative scheme of the MALDI–MSI principle (Gut Y, Boiret M, et al., 2014).
Fig. 1. Representative scheme of the MALDI–MSI principle (Gut Y, Boiret M, et al., 2014).
Detection capability matrix
| Sample Compatibility | Types of Metabolites |
|
|
Workflow
1
Standardized Sample Processing
- Cryosectioning (thickness of 5-20 μm)
- Matrix application optimization
- Histological staining and labeling of mass spectrometry imaging areas
2
Mass Spectrometry Data Acquisition
- Comprehensive full-spectrum scanning in positive and negative ion modes using high-sensitivity MALDI-TOF/Orbitrap platforms
3
In-Depth Data Analysis
- Metabolite annotation
- Differential metabolite screening
- Spatial co-localization network analysis
- Optional AI-driven spatial multi-omics correlation modeling
4
Results Delivery
- Metabolite identification
- Mass spectrometry imaging results
- Relative abundance and spatial distribution
- Detailed analysis report
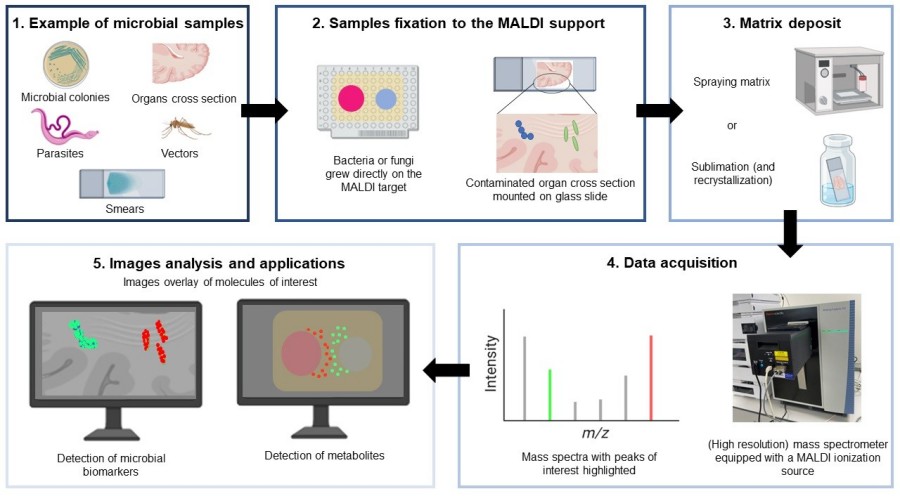 Fig. 2. MALDI imaging MS workflow (Feucherolles M, Frache G. et al., 2022).
Fig. 2. MALDI imaging MS workflow (Feucherolles M, Frache G. et al., 2022).
Features

High Precision and Flexibility
Spatial resolution as low as 5 μm, achieving single-cell detection accuracy, supporting both broad-spectrum analysis and targeted detection.
Integrated Solutions
Combining multi-omics platforms to provide comprehensive data integration for your research.
User-Friendly Data Delivery
Comprehensive customer support and training from sample preparation to result analysis.
What Can You Do with Our Spatial Metabolomics Sloution?
- Cancer Research: Utilized for tumor biomarker screening, distinguishing tumor margins, and studying pharmacokinetics.
- Disease Mechanism Studies: Unveils metabolic heterogeneity in the progression of diseases.
- Pharmacokinetics: Investigates the distribution and concentration of drugs and their metabolites within tissues.
- Botanical Research: Analyzes the spatial distribution of metabolites in plant tissues and studies plant-environment interactions.
- Developmental Biology: Reveals metabolic regulation and mapping during organ and embryo development.
FAQ
Q1: Which types of samples do you support and what are the related requirements?
We support a variety of sample types, including animal tissues (such as brain, tumor, liver, etc.) and plant tissues (such as leaves, stems, etc.). The samples must meet the following requirements: for animal samples, the recommended size is 1 cm × 1 cm × 0.3 cm; for plant leaves, it is 3 cm × 3 cm; and for stems, it is 5 cm × (1~5) cm. Unsupported sample types include unformed tissue blocks, samples with excessively high-water content (≥80%), plant samples with hard shells, and bone samples.
Q2: What types of data analysis do you provide?
We offer a comprehensive range of data analysis services, including metabolite distribution mapping, content information extraction, and differential analysis. Clients can select modular workflows or request fully customized analytical frameworks tailored to specific research objectives.
Q3: What are the advantages of MALDI-MSI technology?
MALDI-MSI technology offers label-free, high-sensitivity, and high-resolution molecular imaging, capable of simultaneously visualizing diverse molecules like lipids and metabolites. It integrates seamlessly with pathological images for contextual insights and is broadly applicable across various research fields, delivering stable and reproducible results with minimal sample preparation.
References
- Gut Y, Boiret M, et al. Application of chemometric algorithms to MALDI mass spectrometry imaging of pharmaceutical tablets. J Pharm Biomed Anal. 2015. 105:91-100.
- Feucherolles M, Frache G. MALDI Mass Spectrometry Imaging: A Potential Game-Changer in a Modern Microbiology. Cells. 2022. 11(23):3900.
Explore Other Options