Fluorescent In Situ Hybridization (FISH) Service
- Overview
- Application
- Service Details
- Case Studies
- FAQs
- Explore Other Options
Fluorescence in situ hybridization (FISH), developed in the 1980s, is a cytogenetic technique using fluorescent probes to bind the chromosome with a high degree of complementarity. It is a powerful and easy method to detect RNA or DNA sequences in cells, tissues, and tumors. This technique is useful for identifying chromosomal abnormalities, gene mapping, characterizing somatic cell hybrids, checking amplified genes and studying the mechanism of rearrangements. RNA FISH is used to measure and localize mRNAs and other transcripts within tissue sections or whole mounts.
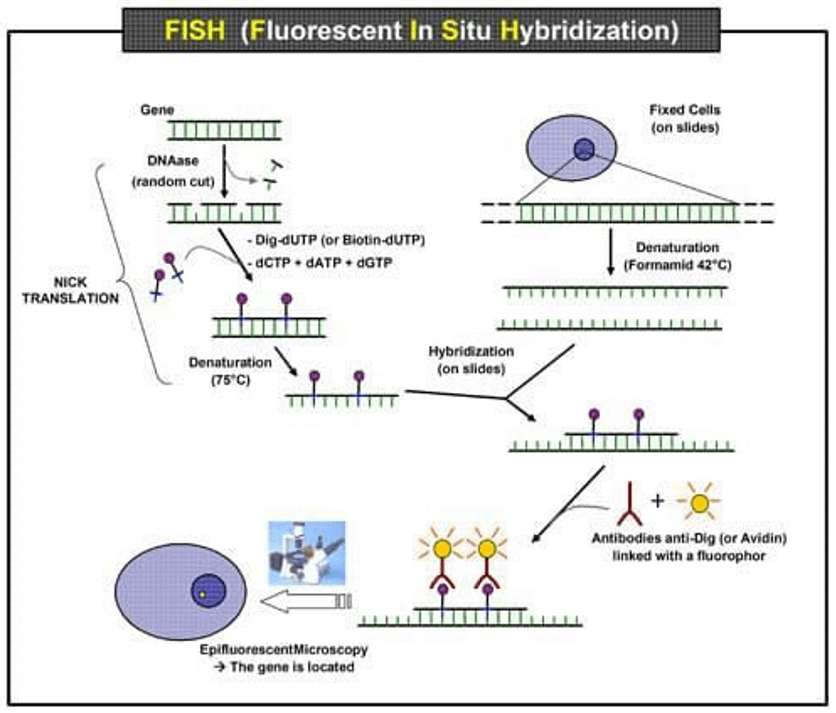 Fig. 1 Scheme of the principle of the FISH experiment to localize a gene in the nucleus.
Fig. 1 Scheme of the principle of the FISH experiment to localize a gene in the nucleus.
FISH is useful, for example, to help a researcher or clinician identify where a particular gene falls within an individual's chromosomes. The first step is to prepare short sequences of single-stranded DNA that match a portion of the gene the researcher is looking for. These are called probes. The next step is to label these probes by attaching one of a number of colors of fluorescent dye. DNA is composed of two strands of complementary molecules that bind to each other like chemical magnets. Since the researchers' probes are single-stranded, they are able to bind to the complementary strand of DNA, wherever it may reside on a person's chromosomes. When a probe binds to a chromosome, its fluorescent tag provides a way for researchers to see its location.
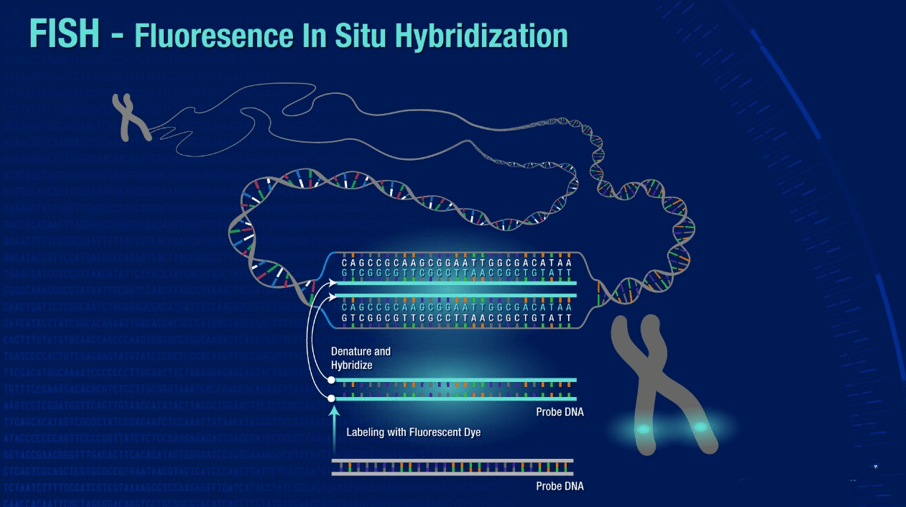
Application of FISH
Scientists use three different types of FISH probes, each of which has a different application:
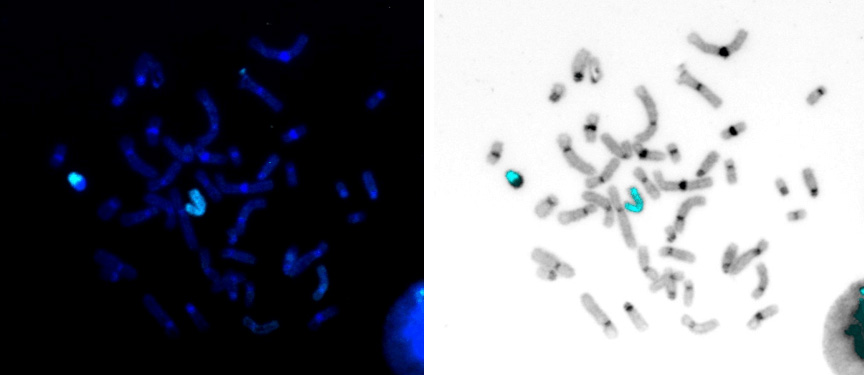 Fig 2. Human chromosome X paint Probe
Fig 2. Human chromosome X paint Probe
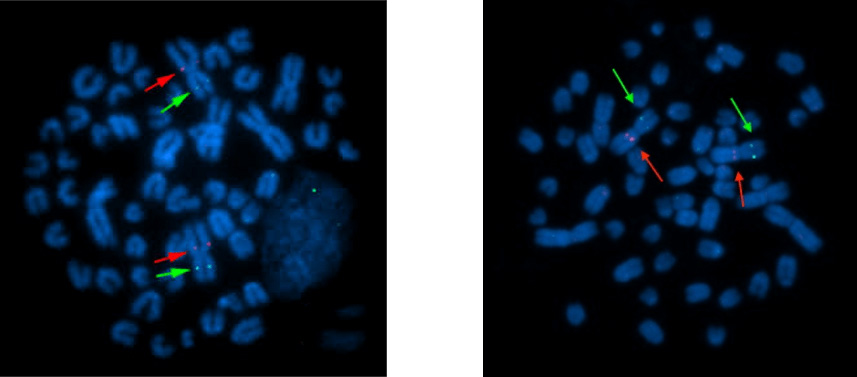 Fig 3. Sheep Specific Gene Probe
Fig 3. Sheep Specific Gene Probe
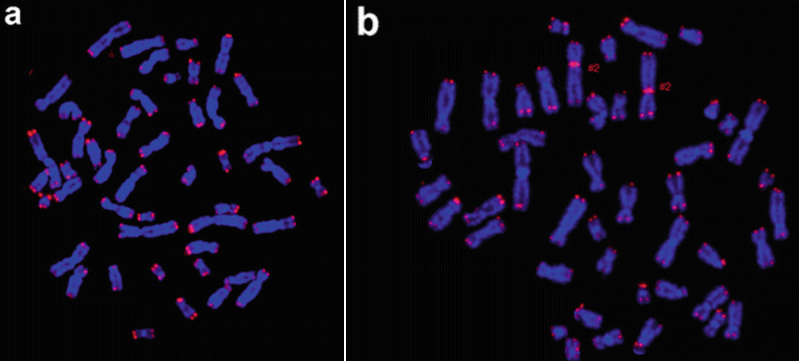 Fig 4. Human Telomere FISH Probe
Fig 4. Human Telomere FISH Probe
Locus Specific Probes bind to a particular region of a chromosome. This type of probe is useful when scientists have isolated a small portion of a gene and want to determine on which chromosome the gene is located, or how many copies of a gene exist within a particular genome.
Alphoid or Centromeric Repeat Probes are generated from repetitive sequences found in the middle of each chromosome. Researchers use these probes to determine whether an individual has the correct number of chromosomes. These probes can also be used in combination with "locus specific probes" to determine whether an individual is missing genetic material from a particular chromosome.
Whole Chromosome Probes are actually collections of smaller probes, each of which binds to a different sequence along the length of a given chromosome. Using multiple probes labeled with a mixture of different fluorescent dyes, scientists are able to label each chromosome in its own unique color. The resulting full-color map of the chromosome is known as a spectral karyotype. Whole chromosome probes are particularly useful for examining chromosomal abnormalities, for example, when a piece of one chromosome is attached to the end of another chromosome.
Creative Bioarray offers a range of different FISH services including:
Metaphase and Interphase FISH (chromosomal assignment and clone ordering)
Fibre-FISH (Chromosome Painting)
3D-FISH (on three-dimensionally preserved nuclei)
FISH on paraffin sections (analysis of archive material)
With combined academic and industrial experiences, we are confident that we will achieve high quality work and guarantee you complete satisfaction. As an important routine technique, FISH could be very time consuming. Creative Bioarray fully understands the details and pitfalls of FISH and histology. We will work side-by-side with you to find the best design to achieve your aims. Our service is efficient and reliable with relatively low cost.
Case Studies
- KRAS Gene Copy Number analysis in A cell line
- Three Gene copy number analysis in FFPE samples
- FISH analysis on Bovine Sperm Slides
KRAS Gene Copy Number analysis in A cell line
Objective: To detect the copy number of the KRAS gene in A cell line.
FISH Probe: KRAS FISH Probe (Cat No.: FON-283)
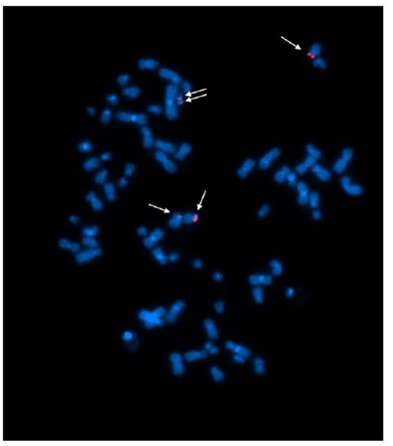 Fig 5. Example of FISH signals in A cell line (#003)
Fig 5. Example of FISH signals in A cell line (#003)
Table 1. The KRAS FISH Signals in 20 Cells
| The Number of KRAS FISH Signals | 3 | 4 | 5 | Total |
| The Number of Cells | 3 | 9 | 8 | 20 |
Three Gene copy number analysis in FFPE samples
Objective: To detect three gene copy number in FFPE samples
FISH Probe: Gene A (7q22.3), Gene B (8q24.3) and Gene C (19q13.2) FISH Probes were created from BAC DNA and directly labeled by nick translation with Cy5-dUTP, Green-dUTP and Cy3-dUTP respectively.
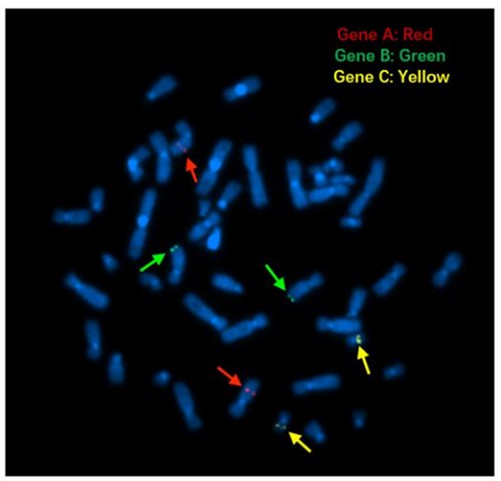 Figure 6. FISH signals on metaphase cells.
Figure 6. FISH signals on metaphase cells.
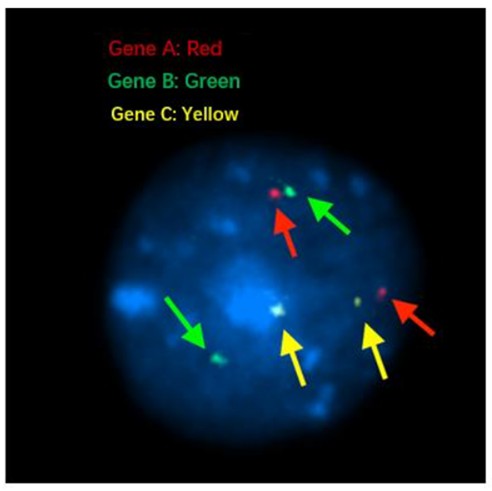 Figure 7. FISH signals on interphase nuclei.
Figure 7. FISH signals on interphase nuclei.
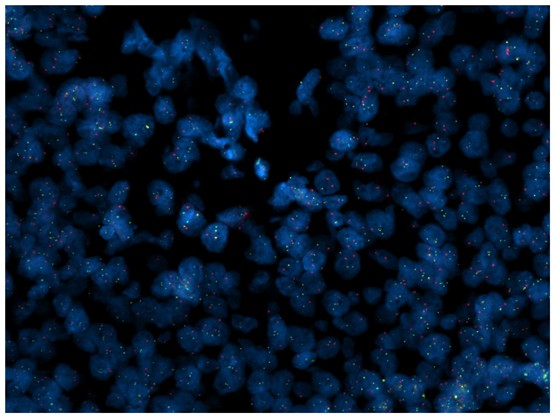 Figure 8. FISH signals on the FFPE sample B.
Figure 8. FISH signals on the FFPE sample B.
Table 2. Summary of FISH signals in the FFPE sample.
| Number of Cells | Number of FISH Signals | ||
| Gene C (yellow) | Gene A (red) | Gene B (green) | |
| 10 | 2 | 2 | 2 |
| 1 | 2 | 2 | 1 |
| 1 | 2 | 2 | 2 |
| 1 | 2 | 1 | 2 |
| 1 | 1 | 1 | 2 |
| 1 | 0 | 2 | 1 |
| 1 | 1 | 1 | 2 |
| 1 | 1 | 2 | 2 |
| 1 | 2 | 3 | 3 |
| 1 | 0 | 3 | 1 |
FISH analysis on Bovine Sperm Slides
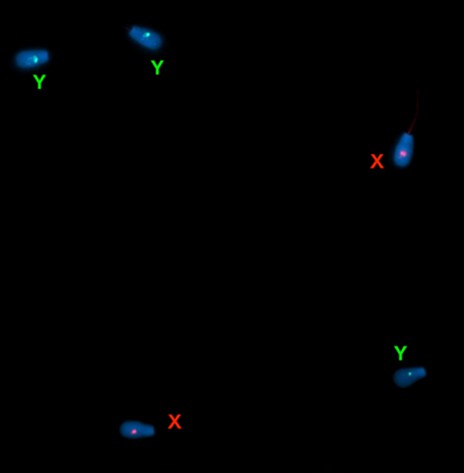 Figure 9. Bovine chromosome XY FISH signals on a bovine sperm slide.
Figure 9. Bovine chromosome XY FISH signals on a bovine sperm slide.
Table 3. Summary of FISH signals in the bovine sperm slide sample.
| File # | X | Y | XY | No Signal |
| 01 | 2 | 0 | 0 | 0 |
| 02 | 2 | 0 | 0 | 0 |
| 03 | 1 | 1 | 0 | 0 |
| 04 | 3 | 0 | 0 | 0 |
| 05 | 2 | 1 | 0 | 0 |
| 06 | 1 | 2 | 0 | 0 |
| 07 | 0 | 2 | 0 | 0 |
| 08 | 2 | 1 | 0 | 0 |
| 09 | 2 | 1 | 0 | 1 |
| 10 | 1 | 1 | 0 | 0 |
FAQs
1. What is Fluorescent In-Situ Hybridization (FISH)?
FISH is a powerful and sophisticated molecular cytogenetic technique that allows for the detection and localization of specific DNA sequences on chromosomes. It utilizes fluorescent probes that bind to target regions of the chromosomes, enabling visualization of genetic material in cells and tissues.
2. What are the applications of FISH?
FISH is widely used in various fields, including:
- Cancer diagnostics and research
- Detection of genetic abnormalities and chromosomal translocations
- Identification of specific chromosome markers
- Genetic mapping and gene expression studies
- Developmental biology and neuroscience research
3. What types of specimens can FISH be performed on?
FISH can be performed on a variety of specimens, including:
- Tissue samples (fresh or fixed)
- Cell cultures
- Blood samples
- Paraffin-embedded tissues
We customize our services based on the specific needs of your project.
4. How do I prepare my samples for FISH?
To ensure the best results, please follow the provided sample collection guidelines. Our team is happy to assist you with any specific preparation requirements for your samples to ensure compatibility with our FISH services.
5. What types of probes do you offer for FISH?
Our service includes a wide selection of custom-designed fluorescent probes tailored to your research needs. Whether you need specific gene probes, chromosome paints, or cancer-related probes, we can create them to suit your specific requirements.
6. Can I request a consultation before placing an order?
Absolutely! Our experts are available for consultations to discuss your specific research needs, project scope, and any questions you may have. We aim to provide personalized support to ensure your success.
7. What quality control measures do you have in place?
At Creative Bioarray, we implement stringent quality control protocols throughout the FISH process to ensure reliable and reproducible results. Our experienced team conducts thorough testing and validation of all probes and sample preparations.
Quotation and ordering
Our customer service representatives are available 24hr a day! We thank you for choosing Creative Bioarray at your preferred Fluorescent In Situ Hybridization (FISH) Service.
Explore Other Options