AP-1060
Cat.No.: CSC-C0596
Species: Homo sapiens (Human)
Source: Bone Marrow
Morphology: single round cells growing in suspension
Culture Properties: suspension
- Specification
- Background
- Scientific Data
- Q & A
- Customer Review
Immunology: CD3 -, CD13 +, CD14 -, CD19 -, CD33 +, HLA-DR -
Viruses: PCR: EBV -, HBV -, HCV -, HIV -, HTLV-I/II -, SMRV -
AP-1060 is a human acute promyelocytic leukemia (APL) cell line established by Sun et al. in 2004 from bone marrow aspirate of a 45-year-old male with clinical resistance to both all-trans retinoic acid (ATRA) and arsenic trioxide (ATO). AP-1060 is a naturally ATRA/ATO dual-resistant model, which is important for understanding the mechanism of APL drug resistance. AP-1060 cells grow in vitro in a suspension form and morphologically remain promyelocytes with large cell bodies, round to indented nuclei, ample cytoplasm with coarse azurophilic granules, and obvious Auer rods.
Molecularly, AP-1060 maintains the classical t(15;17)(q22;q11) translocation which produces the PML-RARA fusion gene. However, p.Pro408Ser mutation (p.Pro900Ser in the fusion protein) was identified in the RARA moiety which blocks the ATRA-induced transcriptional activation. In addition, the cell line was found to acquire a rare secondary t(3;14)(p21.1;q11.2) translocation with the formation of an ETV6-NTRK3 (EN) fusion and homozygous deletion of WT1. These changes are suspected to play a role in drug-resistance acquisition and myeloid transformation potential.
ATRA-Resistant APL Cells are Phenotypically and Mechanically Distinct
Although all-trans retinoic acid (ATRA) is a critical drug for the treatment of acute promyelocytic leukemia (APL), an aggressive form of acute myeloid leukemia, almost 20% of APL patients are resistant to ATRA. As no biomarkers for ATRA resistance are yet available, using mechano-node-pore sensing, a single-cell mechanical phenotyping platform, and patient-derived APL cell lines NB4 (ATRA-sensitive) and AP-1060 (ATRA-resistant), Brain et al. explored whether cell mechanics could be linked to this pathological phenotype.
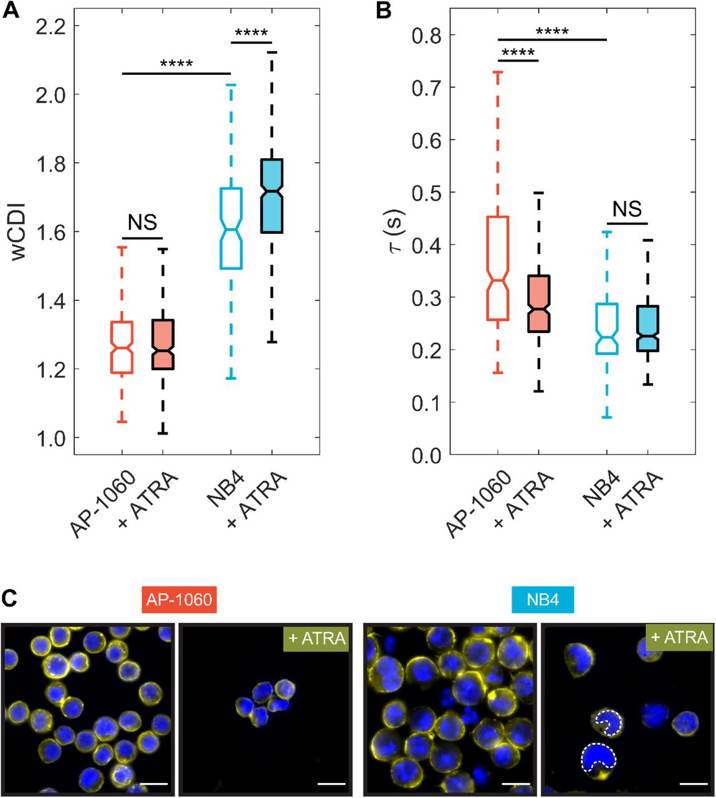
NB4 and AP-1060 cells were cultured in media with ATRA at concentrations ranging from 10 nM to 10 µM and their differentiation was measured. Flow cytometry revealed that NB4 cells were sensitive to ATRA treatment, while AP-1060 cells were mostly resistant. Mechano-phenotyping demonstrated that untreated AP-1060 cells were stiffer than untreated NB4 cells (Fig. 1A). After 4 days of 1 µM ATRA treatment only NB4 cells became more deformable (Fig. 1A). AP-1060 cells had limited maturation and no significant change in deformability. Recovery times were measured to report an apparent viscosity of the cells. AP-1060 cells were more viscous than NB4 cells (Fig. 1B). Treatment with ATRA significantly decreased the recovery time of AP-1060 cells, while there was no significant effect on NB4 cells. Overall, AP-1060 and NB4 cells had different responses to ATRA in both protein expression and mechanical phenotype, with ATRA having differential effects on elasticity (wCDI) and viscosity (recovery time).
Evaluating Sources of Technical Variability in the Mechano-Node-Pore Sensing Pipeline and Their Effect on the Reproducibility of Single-Cell Mechanical Phenotyping
Single-cell mechanical properties offer insights into cellular functions, and various microfluidics-based platforms have been developed for high-throughput testing. However, the technical variability and reproducibility of these platforms have not been fully characterized. Brain et al. evaluated the repeatability of mechano-node-pore sensing, a single-cell mechanical phenotyping platform, by assessing the impact of device-to-device variability and semi-manual data processing on measurement accuracy.
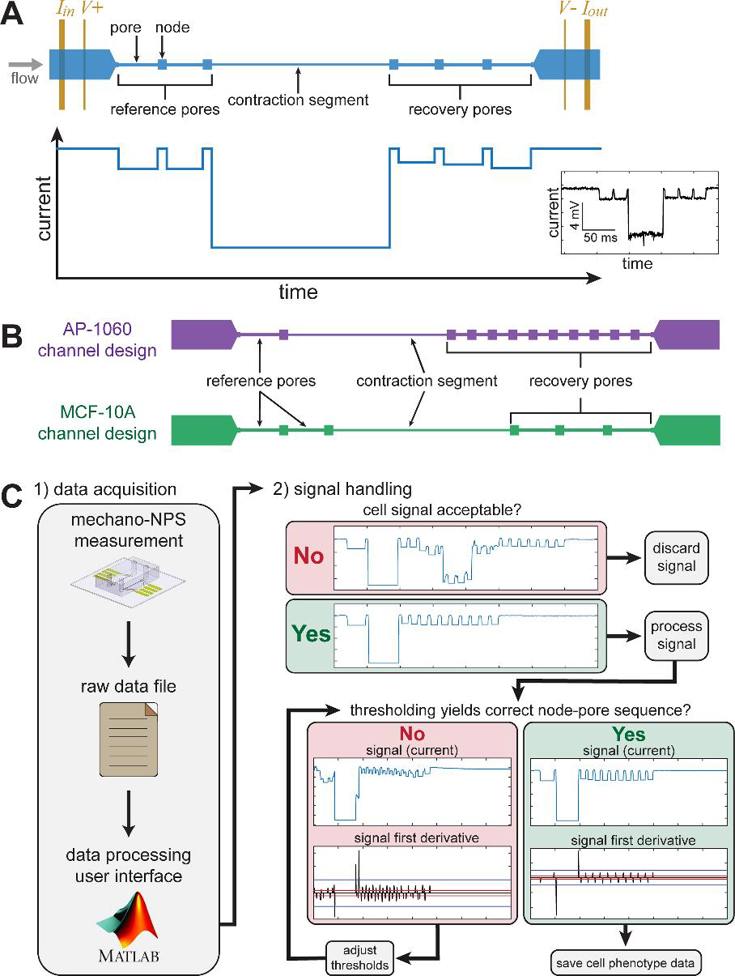
In node-pore sensing (NPS), a microfluidic channel is divided into wider "nodes" and narrower "pores." As cells are flowed through the device, characteristic current pulses are measured in a four-terminal configuration (Fig. 2A). The magnitude of these pulses is determined by the size of the cell and the transit time. In mechano-NPS, cells flow through several reference pores, a contraction segment, and recovery pores. The reference pores measure the cell's initial diameter and velocity, the contraction segment measures the cell's resistance to deformation, and the recovery pores measure the time taken to relax to the original size and shape. Each mechano-NPS device is optimized for a specific cell size. To evaluate the reproducibility, they re-used the same device designs from Kim et al. and Li et al. to measure MCF-10A and AP-1060 cells, respectively (Fig. 2B). The elastic properties of the cells are quantified with a whole-cell deformability index (wCDI), while the viscoelastic behavior is quantified with a recovery time. They quantified device-to-device variability and the impacts of data processing on the reproducibility by analyzing AP-1060 cells (Fig. 2C). They also examined the overall reproducibility in two labs using identical devices and MCF-10A cells.
Ask a Question
Write your own review
- You May Also Need
- Adipose Tissue-Derived Stem Cells
- Human Neurons
- Mouse Probe
- Whole Chromosome Painting Probes
- Hepatic Cells
- Renal Cells
- In Vitro ADME Kits
- Tissue Microarray
- Tissue Blocks
- Tissue Sections
- FFPE Cell Pellet
- Probe
- Centromere Probes
- Telomere Probes
- Satellite Enumeration Probes
- Subtelomere Specific Probes
- Bacterial Probes
- ISH/FISH Probes
- Exosome Isolation Kit
- Human Adult Stem Cells
- Mouse Stem Cells
- iPSCs
- Mouse Embryonic Stem Cells
- iPSC Differentiation Kits
- Mesenchymal Stem Cells
- Immortalized Human Cells
- Immortalized Murine Cells
- Cell Immortalization Kit
- Adipose Cells
- Cardiac Cells
- Dermal Cells
- Epidermal Cells
- Peripheral Blood Mononuclear Cells
- Umbilical Cord Cells
- Monkey Primary Cells
- Mouse Primary Cells
- Breast Tumor Cells
- Colorectal Tumor Cells
- Esophageal Tumor Cells
- Lung Tumor Cells
- Leukemia/Lymphoma/Myeloma Cells
- Ovarian Tumor Cells
- Pancreatic Tumor Cells
- Mouse Tumor Cells