GUIDELINE
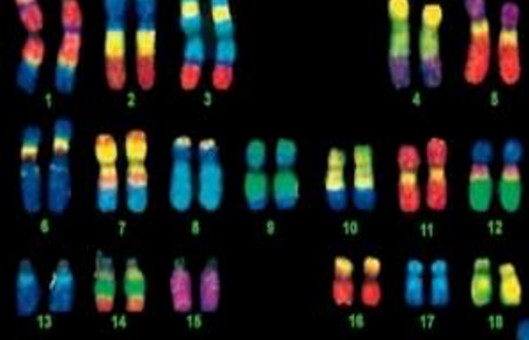
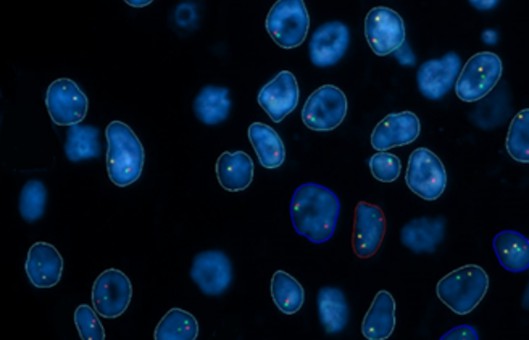
- Fluorescence in situ hybridization (FISH) began with the combination of traditional cytogenetic and DNA techniques.
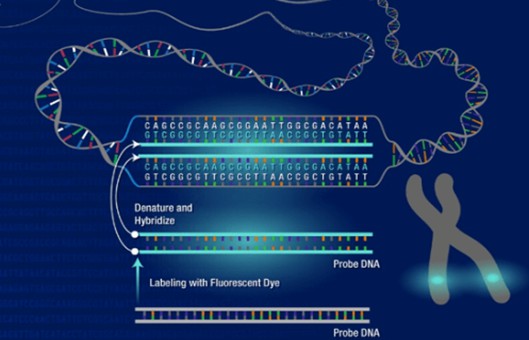
- It is based on the Southern blot principle, where a known nucleic acid molecule indirectly labeled with a semi-antigen such as biotin, digoxin or directly labeled with fluorescein is used as a probe. The probe and target sequence double-stranded DNA is denatured and then hybridized. The complementary heterologous single-stranded DNA molecules are annealed at a suitable temperature and ionic strength to form stable heterologous double-stranded DNA, the semi-antigens are displayed by fluorescently labeled affinity or anti-digoxin antibodies, and the hybridization signal is observed by fluorescence microscopy.
METHODS
Notch translation marker probes
- The notch panning system for 1μg DNA is 0.1 mmol/L dTTP 6.5 μl, 0.1 mmol/L dNTP 10 μl, 10×Nick Translation buffer 5 μl, 1 mmol/L DIG-11-dUTP /Biotin-16-dUTP 0.5 μl, Nick Translation Enzyme 10 μl, DNA (1μg) X μl, and H2O 18-X μl. For plasmids ≤10kb, react at 15°C for 3.5 hours; for BAC/PAC, react at 15°C for 9 hours.
- Place the EP tube on ice, take 5 μl of it, and heat it at 70°C for 5 minutes, then perform 2% agarose gel electrophoresis to observe the size of the labeled probe, the suitable size of the single sequence probe is 200-600 bp; if the fragment size is large, extend the reaction at 15°C for 10-30 minutes, then repeat the gel electrophoresis until the suitable size of the probe is obtained.
Precipitation and denaturation of probes
- Composition of the probe mixture. For BAC/PAC probes, DNA probe 5 μl, Human Cot-1 DNA 3 μl, Salmon Sperm DNA 0.5 μl, H2O 1.5 μl. For mitotic probes, DNA probe 5 μl, Salmon Sperm DNA 0.5 μl, H2O 4.5 μl.
- Precipitation of DNA. The probe mixture was mixed with 1 μl (V/10) 3M sodium acetate (pH 5.2) and 27.5 μl (2.5V) anhydrous ethanol (lyophilized at -20°C), precipitated at -80°C for 30 min (or -20°C overnight), and centrifuged at 14,000 g at 4°C for 30 min to precipitate DNA.
- Wash the precipitate. Carefully discard the supernatant, wash once with 70% ethanol, and centrifuge again at 14,000 g for 15 min at 4°C; carefully discard the supernatant and air-dry the DNA precipitate in a medium temperature water bath at 45-50°C for 10-15 min.
- Dissolve the probe. Add 5 μl of pre-warmed deionized formamide (pH 7.0) at 37°C, centrifuge for a short time and shake at 37°C for 30 min to fully dissolve the DNA; then add 5 μl of pre-warmed Master Mix at 37°C, centrifuge for a short time and shake at 37°C for 15-30 min.
- Denaturation and pre-hybridization of probes. Short centrifugation followed by denaturing probes in an 80°C water bath for 10 min and an ice bath for 5 min; short centrifugation and pre-hybridization in a 37°C water bath for 30-60 min; unique sequence probes (e.g. cDNA) and repeat sequence probes (e.g. α-satellite DNA) probes, no pre-hybridization is required.
- Hybridize at 37°C overnight in a humidity chamber.
Creative Bioarray Relevant Recommendations
- Creative Bioarray provides the most comprehensive list of FISH probes for the rapid identification of a wide range of chromosomal aberrations across the genome. We continue to expand the scope of the FISH probes to meet the customer's research needs- including the introduction of Customized FISH Probes.
NOTES
- When most of the ethanol evaporates, the white DNA precipitate becomes translucent; be sure to remove the ethanol thoroughly, otherwise, tiny precipitates will be generated after the addition of Master Mix, resulting in high background.
- The DNA precipitate can also be dissolved in TE buffer 1.1 μl and diluted by adding Vysis probe buffer 4.4 μl, mixed and centrifuged instantaneously, and shaken for 15-30 min at 37°C.
RELATED PRODUCTS & SERVICES