ImmunoFISH: Integrates FISH and IL for Dual Detection
The ability to visualize specific DNA or RNA sequences within their natural environment has, for many years, been made possible by Fluorescent In Situ Hybridization (FISH). Using this method researchers can now observe genomic aberrations with unparalleled spatial resolution. Similarly, immunolabelling (IL) has empowered researchers to map protein localization and expression with high specificity. Yet, when used in isolation, each technique presents a fundamental limitation: they offer only a single-layer snapshot. FISH reveals the "genetic blueprint," while immunostaining shows the "functional machinery," but they cannot connect the two within the very same cell.
This is a major hurdle in the context of many intricate biological questions. How is a specific chromosomal translocation responsible for the altered protein signaling driving the oncogenic state? Is a key transcription factor targeting the gene it is known to regulate during cellular differentiation? The demand for correlative analysis of DNA, RNA, and protein information, in particular for studies in cancer research, cell line characterization, and developmental biology is rapidly increasing.
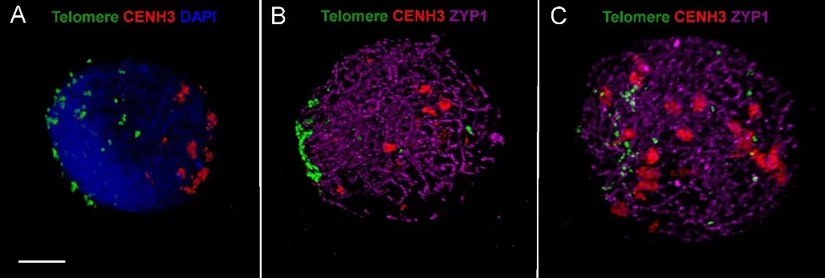
So, What Exactly is ImmunoFISH?
Simply put, ImmunoFISH is a multi-methodological workflow that sequentially performs IL and FISH on the same biological sample. This leads to the simultaneous detection of selected proteins (via antibodies conjugated to fluorophores) and selected DNA or RNA sequences (via nucleic acid probes labelled with fluorophores of a different colour) in single nuclei/cells. In other words, it merges genomics and proteomics in one image, by enabling the direct correlation of genetic information with its functional protein output.
The Core Principle Behind the Technique
The key to success with ImmunoFISH is a well-balanced protocol that preserves the antigenic epitopes (for antibody binding) and nucleic acid structure (for probe hybridization). Protocols may vary, but the basic workflow involves these essential steps:
- Cell Fixation and Permeabilization: Fix cells or tissue sections (typically with paraformaldehyde) to preserve cellular architecture and immobilize all molecules in place. Permeabilize the cells to allow antibodies and probes access.
- Immunolabelling (First): The immunostaining step should (practically always) be performed first. The harsh denaturing conditions of FISH (high temperature, formamide) will destroy the 3D structure of proteins and make them unrecognizable to antibodies. Primary antibodies are applied to bind to the target protein, then fluorescently-conjugated secondary antibodies.
- Fixation (Optional but Recommended): A "locking in" fixation step is typically inserted after immunolabelling to "lock" the antibodies in place and prevent them from being removed or degraded during the subsequent rigorous FISH procedure.
- Fluorescent In Situ Hybridization (Second): Next, the standard FISH protocol is performed. This involves denaturing the chromosomal DNA (if you are detecting DNA targets) to make it single-stranded, then applying labeled nucleic acid probes that will find their complementary sequence to hybridize to.
- Counterstaining and Imaging: Finally, a general nuclear counterstain such as DAPI is added. The sample is then imaged on a fluorescence microscope with appropriate filter sets to differentiate between the emission spectra of the DAPI, the immunofluorescence signal and the FISH signal.
By following these steps in order, it is possible to create a multi-parametric snapshot of a single cell.
Major Applications of ImmunoFISH
Cancer Cytogenetics
ImmunoFISH is increasingly used in oncology research to correlate chromosomal translocations or amplifications with the activation of oncogenic proteins.
For instance, in breast cancer, it can link HER2 gene amplification with HER2 protein overexpression, aiding in the validation of HER2 status and its downstream signaling pathways.
This concurrent detection facilitates tumor classification, prognosis assessment, and targeted therapy development.
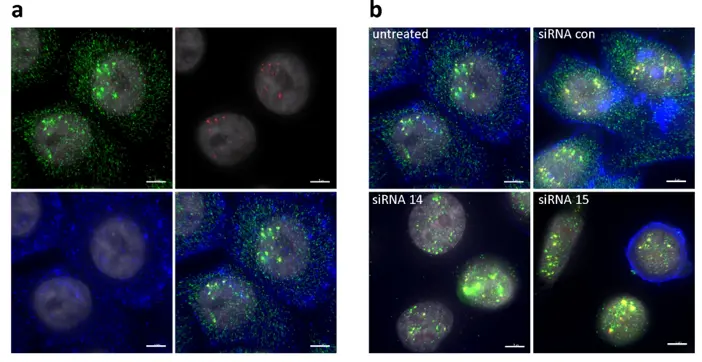
Stem Cell and Developmental Biology
Cell differentiation is marked by a well-timed regulation between transcription and translation to commit a cell to a specific lineage.
ImmunoFISH can be used to study the dynamic process of cellular differentiation by observing gene activation events in concert with lineage-specific proteins.
It is particularly useful in research related to embryogenesis, tissue regeneration, and stem cell plasticity.
Viral Integration Studies
ImmunoFISH can be utilized to visualize viral genomes integrated into the host DNA in conjunction with viral or host response proteins.
This approach helps in understanding the mechanisms of viral latency, reactivation, and oncogenic transformation, which is crucial in viruses like HPV, HBV, and HIV.
Drug Discovery and Mechanistic Studies
ImmunoFISH can be used as a valuable assay during drug discovery to study the impact of therapeutic agents on gene regulation and consequent protein expression.
Monitoring both genetic and phenotypic responses simultaneously in the same cell population helps to understand drug efficacy, off-target effects, and pathway modulation.
Cell Division and Chromosomal Dynamics
ImmunoFISH has an important application in the study of cell cycle progression and mitosis.
It allows the visualization of centromeric DNA and the associated kinetochore proteins during cell division, providing insights into the molecular events that maintain genomic stability and chromosomal segregation fidelity.
Why ImmunoFISH Outperforms Conventional FISH
| Feature | Traditional FISH | ImmunoFISH |
|---|---|---|
| Detection Targets | DNA/RNA sequences | DNA/RNA + proteins |
| Information Depth | Genomic | Integrated genotype-phenotype |
| Spatial Resolution | High | High (with additional molecular context) |
| Applications | Cytogenetics, mutation detection | Cancer, developmental biology, diagnostics |
| Data Integration | Limited | Comprehensive - from genes to function |
Creative Bioarray Relevant Recommendations
| Products & Services | Description |
|---|---|
| ImmunoFISH Analysis (FISH+IHC) | Creative Bioarray offers the ImmunoFISH analysis from the assay development, validation and final testing and analysis services. |
| Fluorescent In Situ Hybridization (FISH) Service | Creative Bioarray offers a full line of Fluorescent in situ Hybridization (FISH) services, from standardized testing of validated assays to custom development of new assays. |
| Immunohistochemistry (IHC), Immunofluorescence (IF) Service | Creative Bioarray offers a comprehensive IHC service from project design, marker selection to image completion and data analysis. |
References
- Sepsi A, Fábián A, et al. ImmunoFISH: Simultaneous Visualisation of Proteins and DNA Sequences Gives Insight into Meiotic Processes in Nuclei of Grasses. Front Plant Sci. 2018. 9:1193.
- Kwon S, Chin K, et al. Quantitative, in situ analysis of mRNAs and proteins with subcellular resolution. Sci Rep 7, 16459 (2017).