Cell Immortalization Step by Step
Primary cells typically stop dividing after 40-60 passages, a result of replicative senescence (the Hayflick limit). This severely constrains the length and reproducibility of long-term experiments. Immortalized cell lines, in contrast, are cells which, as a result of genetic manipulation, are capable of infinite replication. This is because of the neutralization of the senescence pathways in the cell, making these cell lines grow indefinitely and consistently. Immortal cell lines, therefore, provide an experimentally reliable, nearly limitless source of identical cells.
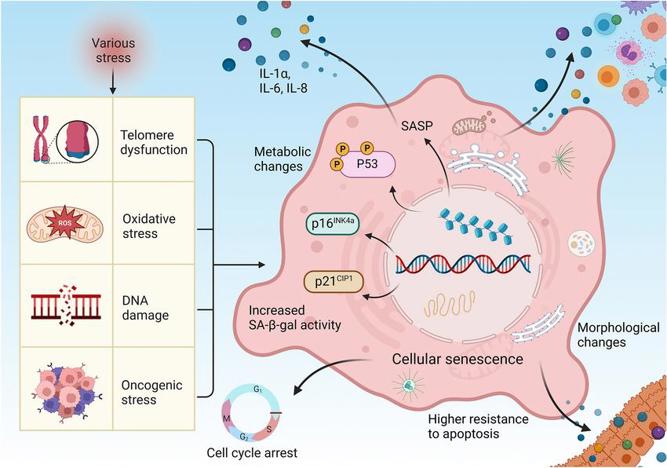
Cellular senescence mechanism
To create an immortal cell line, one must first understand the mechanisms of cellular aging. Cellular senescence is primarily driven by three core molecular pathways:
- Telomere Shortening: Linear chromosomes are protected by specialized DNA-protein caps at their ends known as telomeres. Due to the "end-replication problem", a cell's telomeres become shorter with every cell division. When the telomeres are short enough to be mistaken for damaged DNA, they trigger the senescence program.
- Tumor Suppressor Pathway Activation: The two key tumor suppressor proteins involved in the senescence program are the p53 and pRb (retinoblastoma) family of proteins. These proteins can be described as two molecularly distinct checkpoints that respond to DNA damage and/or telomere dysfunction by triggering cell cycle arrest or apoptosis. Activation of the tumor suppressor pathway is the primary enforcer of the Hayflick limit.
- Accumulation of DNA damage: Cumulative DNA damage over time due to internal and external factors is also known to induce senescence programs.
Comparison of Cell Immortalization Methods
| Method | Main Mechanism | Applicable Cell Types | Typical Advantages | Main Limitations |
|---|---|---|---|---|
| Viral Oncogenes (SV40 Large T, HPV E6/E7, EBV EBNA1/LMP1) | Inhibit tumor suppressor pathways such as p53 and Rb, thereby removing senescence checkpoints | Adherent cells, epithelial cells, B cells, etc. | High transfection efficiency and rapid immortalization; applicable across multiple species | May cause genomic instability and phenotypic alterations; carries potential oncogenic risk |
| Telomerase Activation (hTERT) | Maintains telomere length to prevent replicative senescence | Most human primary cells (fibroblasts, epithelial cells, stem cells, etc.) | Genomic stability is relatively preserved; retains original phenotype; suitable for long-term culture models | Inefficient when used alone in certain cell types (e.g., some hematopoietic cells); often requires additional factors |
| Cell Cycle Regulatory Genes (CDK4 R24C + Cyclin D1, p16 siRNA, p53 siRNA, MYC) | Disrupt the p16-CDK4/6-Rb axis or suppress p53 to drive continuous proliferation | Cells requiring bypass of Rb/p16 inactivation, such as human hepatocytes or tissue cells derived from embryonic stem cells | Significantly increases immortalization efficiency when combined with hTERT; introduces relatively controllable genomic perturbation | CDK4 R24C alone can extend lifespan but may not fully immortalize; requires Cyclin D1 or hTERT co-expression for stable propagation |
| Combination Strategy (hTERT + CDK4 R24C + Cyclin D1, etc.) | Simultaneously extends telomeres and releases cell cycle inhibition, achieving efficient immortalization with high phenotypic fidelity | Primary cells unresponsive to single factors, such as hepatocytes, pancreatic cells, and keratinocytes | High immortalization efficiency (>80%); maintains morphology and function; suitable for pharmacokinetic and metabolic studies | Successfully established in multiple mammalian species (human, bovine, porcine, dolphin) |
| Chemical/Physical Induction (Radiation, Carcinogenic Chemicals, Chronic Hypoxia) | Induces DNA damage or stress responses, allowing selective escape from senescence | Mainly used for susceptible cells such as mouse or rat embryonic fibroblasts | Simple operation without genetic transfection | Low immortalization efficiency; often associated with extensive genomic mutations and unstable phenotypes |
| Viral Oncogenes (SV40 Large T, HPV E6/E7, EBV EBNA1/LMP1) | Inhibit tumor suppressor pathways such as p53 and Rb, thereby removing senescence checkpoints | Adherent cells, epithelial cells, B cells, etc. | High transfection efficiency and rapid immortalization; applicable across multiple species | May cause genomic instability and phenotypic alterations; carries potential oncogenic risk |
Simplified Workflow for Immortalized Cell Line Construction
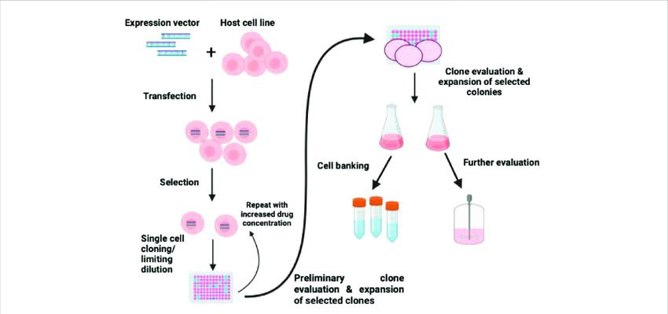
- Isolation and Primary Cell Culture:
Acquisition of high-quality, early-passage primary cells from the tissue of interest. Special focus on quality control. - Vector Construction:
Cloning of the gene of interest for immortalization (hTERT, SV40 LT or other) into an expression vector (usually lentiviral or retroviral) that is typically paired with a selection marker (e.g. antibiotic resistance). - Gene Delivery (Transduction/Transfection):
Introduction of the immortalization construct into the primary cells, most often through highly efficient lentiviral transduction. - Selection and Screening:
Application of antibiotic selection pressure to isolate the successfully immortalized cells that have stably integrated the exogenous DNA. - Long-Term Culture and Expansion:
Culturing the selected cell until they have reached the expected Hayflick limit and show sustained, indefinite proliferation. - Characterization and Validation (Quality Control):
Validation testing to confirm that cells are immortalized (e.g. passage over multiple months, Telomerase activity assay) and that they retain the phenotype, genotype, morphology, and function of the parental primary cells (e.g. flow cytometry, western blot, STR profiling).
Key Considerations
Vector Selection
Choose viral or plasmid vectors that have undergone safety evaluation. Avoid maintaining antibiotic-resistance-containing vectors in non-selective media for extended periods to prevent unintended spread of resistance genes.
Antibiotic Selection Concentration
Determine the minimum lethal concentration (MLC) through gradient testing to identify the optimal selection pressure. Excessively high concentrations may cause widespread cell death, while overly low concentrations can allow untransduced cells to persist.
Verification of Gene Expression
Ensure the stable expression of the immortalization gene by performing qPCR, Western blot or alternative assays. Antibiotic resistance alone is insufficient evidence of successful immortalization.
Cell Morphology and Proliferation
Immortalized cell lines often exhibit distinct compact and uniform morphology and loss of contact inhibition and will proliferate indefinitely. Note passage number to ensure that cells are indeed past the Hayflick limit.
Chromosomal Stability and Mycoplasma Testing
Immortalization may be accompanied by chromosomal instability; therefore, perform periodic karyotype analysis. Mycoplasma contamination can significantly alter cellular behavior and experimental results-routine screening is essential.
Biosafety
When using oncogenic viruses (e.g., SV40, EBV), conduct all procedures in a BSL-2 laboratory. Maintain detailed experimental records, ensure complete inactivation of liquid waste, and comply with all institutional biosafety and ethical regulations.
Creative Bioarray Relevant Recommendations
| Products & Services | Description |
|---|---|
| Cell Immortalization Service | Reliable and risk-free cell immortalization from fibroblasts, epithelial cells, muscle cells from human, rat, mouse, porcine and bovine. |
| Immortalized Cell Line Engineering Service | Our project managers and subject matter experts will work with you to produce immortalized cells tailored to your requirements. |
References
- You L, Nepovimova E, et al. Mycotoxins and cellular senescence: the impact of oxidative stress, hypoxia, and immunosuppression. Arch Toxicol. 2023. 97(2):393-404.
- Maqsood M I., Matin M M, et al. Immortality of cell lines: Challenges and advantages of establishment. Cell Biol. Int. 2013. 37, 1038-1045.