UGT Inhibition (and Other Non-CYP Enzymes)
- Service Details
- Workflow
- Features
- FAQ
- Explore Other Options
Although cytochrome P450 (CYP) enzymes are the most studied and largely account for metabolic enzyme activity, non-CYP enzymes are also critical to the metabolism, clearance, and drug-drug interactions (DDI) of drugs. These enzymes are widely distributed in the liver, kidneys, intestines, and lung tissues, encompassing Phase I and Phase II reactions of drug metabolism, and participate in various metabolic pathways including oxidation, reduction, hydrolysis, and conjugation.
Non-CYP enzymes are highly specific for their substrates, and their inhibition can cause reduced capacity for drug metabolism, leading to increased blood drug levels, and possibly toxic or efficacy-altering reactions. As the role of non-CYP enzymes in clinical pharmacokinetic research is increasingly recognized, thorough evaluations of non-CYP enzyme inhibition potential during the early stages of drug development have become an essential part of new drug application submissions.
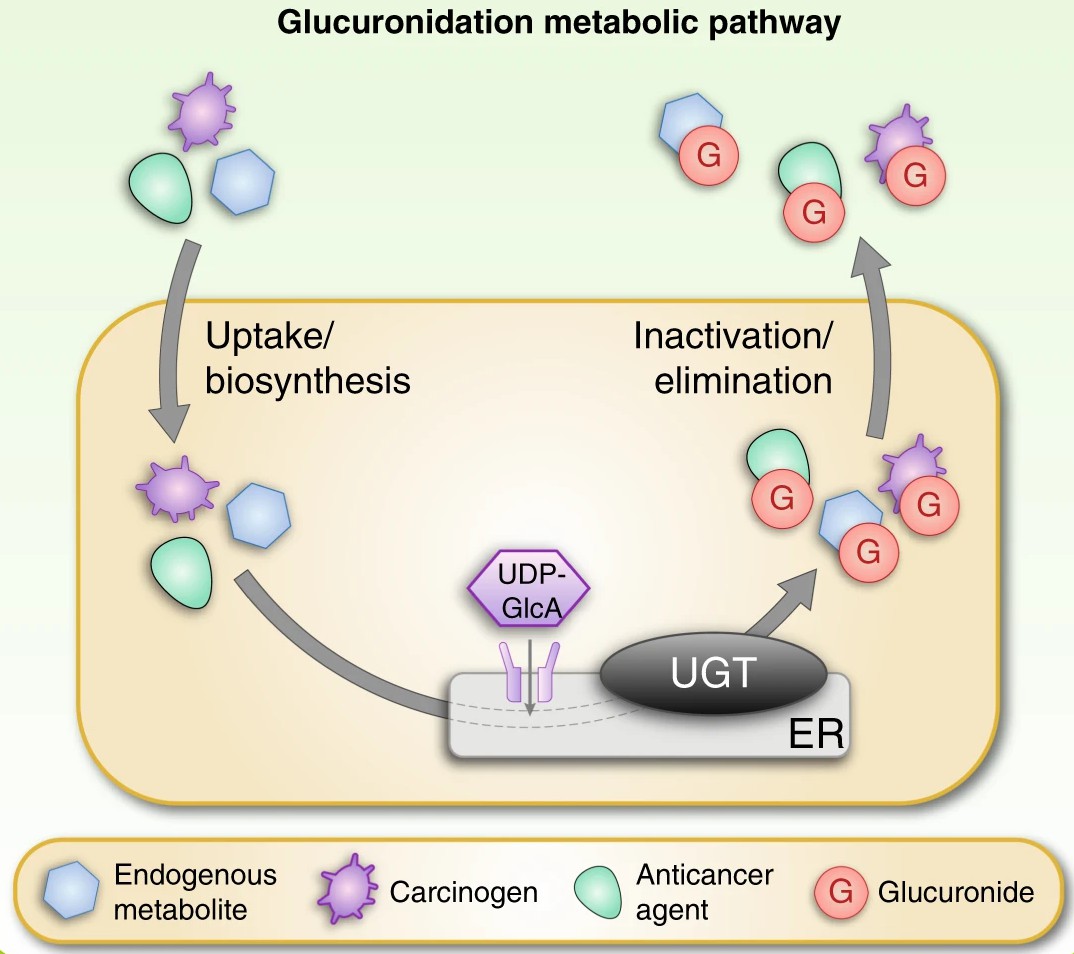 Fig. 1. Schematic overview of the glucuronidation reaction catalysed by UGT enzymes (Allain E P, Rouleau M, et al., 2020).
Fig. 1. Schematic overview of the glucuronidation reaction catalysed by UGT enzymes (Allain E P, Rouleau M, et al., 2020).
Our Comprehensive Enzyme Inhibition Assay Services
Detection Range – Precisely Covering Key Metabolic Enzymes
| Enzyme Category | Representative Subtypes | Main Function | Recommended Detection System |
| UGTs | UGT1A1, UGT1A3, UGT1A4, UGT1A6, UGT1A9, UGT2B7, UGT2B15 | Catalyze glucuronidation reactions | Liver microsomes, Recombinant UGT enzymes |
| SULTs | SULT1A1, SULT1E1 | Catalyze sulfation reactions | Liver S9 fractions, Recombinant SULT enzymes |
| FMO | FMO1, FMO3, FMO5 | Oxidize sulfur/nitrogen-containing drugs | Recombinant FMO enzymes, Liver microsomes |
| XO | XO (Xanthine Oxidase) | Catalyze the oxidation of purine drugs | Recombinant XO enzymes |
| AO | AOX1 (Aldehyde Oxidase 1) | Metabolize anticancer and antiviral drugs | Liver cytosol |
| CES | CES1, CES2 | Hydrolyze prodrugs to generate active metabolites | Liver/intestinal microsomes, Recombinant CES enzymes |
Protocol
| Item | Content |
| Detection Systems | Recombinant enzymes, Liver microsomes, Liver S9 fractions, Liver cytosol |
| Reaction Conditions | 37°C, pH 7.4, Reaction time 15-60 minutes (adjust based on enzyme type) |
| Concentration Gradient | 5-7 concentration gradients covering the expected IC₅₀ range |
| Positive/Negative Controls | Includes FDA-recommended inhibitors (e.g., Atazanavir, Diclofenac) |
| Detection Methods | LC-MS/MS, HPLC, or Spectrophotometry (chosen based on substrate properties) |
Workflow
Client submits compound information and detection requirements.
Our professional team formulates a customized experimental plan.
Experimental implementation (selection of recombinant enzymes or in vivo system according to the requirements).
Data collection and analysis (including comparison with positive controls).
Submit the final experimental report and raw data package.
Features

Professional Team
Experienced pharmacologists and biochemists form our team to guarantee the design and interpretation of experiments and results.

Comprehensive Coverage
We provide multiple detection services for UGTs and non-CYP enzymes to meet different drug development needs.

Efficient and Accurate
We use the latest experimental methods and equipment to ensure the detection results are reliable and repeatable.

Personalized Services
We offer personalized experimental plans based on clients' needs and flexible detection options.

Data Support
We provide multi-metric analysis (IC₅₀, Ki, types of inhibition, etc.) and follow-up consulting.
FAQ
1. What should I do if the tested compound shows strong inhibitory effects in the UGT inhibition assay?
If the UGT inhibition assay indicates potent inhibitory effects, we recommend further evaluating the compound's in vivo pharmacokinetic properties and potential drug-drug interaction risks. We also offer relevant consulting services to support these assessments.
2. Can UGT inhibition detection be conducted alongside CYP inhibition?
Yes, we also recommend performing a complete metabolic enzyme inhibition profile analysis to enable a comprehensive assessment of DDI risk.
3. Is it possible to detect only a specific UGT enzyme (like UGT1A1)?
Certainly. We support flexible combinations of single enzyme types or multiple enzyme types for detection according to the needs of different research stages.
Quotation and Ordering
For complete experimental plans, pricing, or method validation support, please feel free to contact our technical team. We are committed to providing professional, flexible, and high-quality technical support for your drug metabolism research.
Reference
- Allain E P, Rouleau M, et al. Emerging roles for UDP-glucuronosyltransferases in drug resistance and cancer progression. Br J Cancer. 2020. 122, 1277–1287.
Explore Other Options