An Exosome-Related RNA Risk Model for Predicting Breast Cancer
Scientific Report. 2022 Dec 24; 12(1): 22322
Authors: Qiu P, Guo Q, Lin J, Pan K, Chen J, Ding M.
INTRODUCTION
- Breast cancer (BC) is one of the most frequent malignancies among women worldwide. Accumulating evidence indicates that long non-coding RNA (lncRNA) may affect BC progression.
- Exosomes, a class of small membrane vesicles, have been reported to promote tumor progression through transporting proteins, mRNAs, lncRNAs and some other small molecules. However, the interaction between exosome-related lncRNAs and the microenvironment of malignancies is unclear.
METHODS
- Initially, a total of 3158 lncRNAs were obtained by analyzing the RNA-seq data of 828 BC samples and 112 normal breast samples from TCGA. Besides, the correspond clinical information of 828 patients were extracted from TCGA. A combination of several methods was employed to establish a 15-lncRNA risk model and investigated the potential mechanisms of these lncRNAs affect the prognostic outcomes of BC.
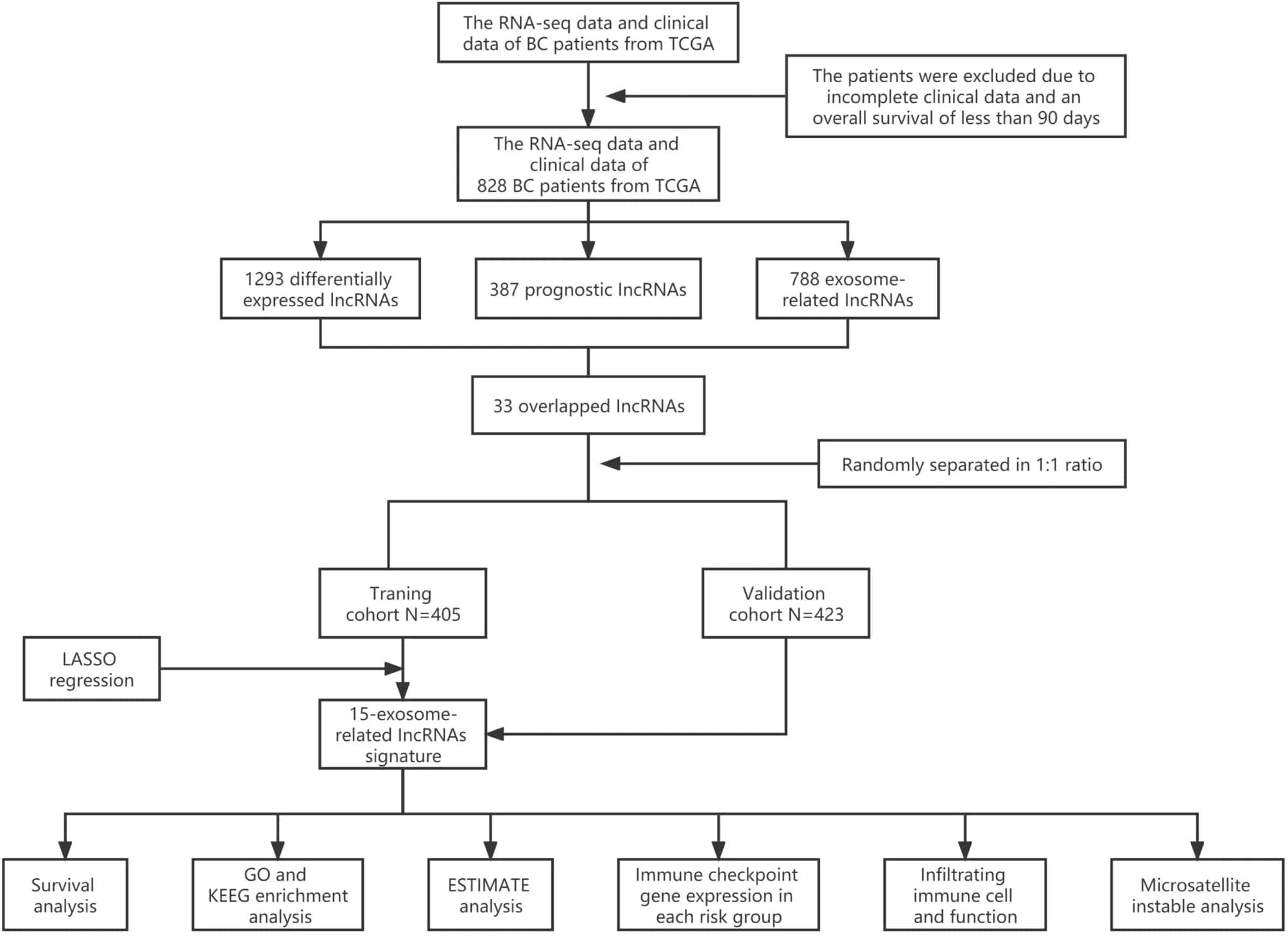 Fig. 1 Analysis flow chart.
Fig. 1 Analysis flow chart.
- The risk score of each patient on the basis of this risk model was calculated through the normalized exosomal lncRNAs expression levels and correspond coefficients. The survival analysis and the time-dependent ROC curve analysis were conducted in validation cohort. As consequence, underwent univariate and multivariate COX regression analysis, the risk score was testified as the independent prognostic element for BC patients.
- A lncRNA-mRNA co-expression network was established included 24 lncRNA-mRNA pairs to investigate the potential roles of the 15 exosome-related lncRNAs in BC. GO and KEGG analysis were adopted to identify the biological functions and signaling pathways associated with the exosome-related risk score. To analyze the tumor microenvironment (TME) landscape and the overall degree of immune infiltration, we calculated the ESTIMATE score of each sample by ESTIMATE algorithm.
- To investigate the potential role of the exosome-associated-lncRNA risk signature in prediction of immunotherapy response, immune checkpoint blockade key molecules, microsatellite instable (MSI) in tumor tissue were further analyzed.
RESULTS
- A total of 15 exosome-related lncRNAs (MEF2C-AS1, SOCS3-DT, LINC01711, MRPL20-DT, LINC00702, MAL2-AS1, USP30-AS1, WASHC5-AS1, TBL1XR1-AS1, LINC02463, LINC02037, ZBTB20-AS1, BTBD9-AS1, SLC24A3-AS1, CACNA1C-IT3) were obtained for follow study.
- The patients were separated to high-risk and low-risk cohorts on the basis of the median risk score, patients with high-risk score are manifested worse survival rates in validation set via the Kaplan-Meier curves. Remarkably, the risk score had been validated robust predictive value for BC survival, presented in the time-dependent ROC analysis. Hence, a 15-lncRNA signature was successfully established to predict prognostic outcomes of BC patients.
- The outcomes of univariate Cox regression analysis revealed that the risk score was an independent prognostic factor for BC patients in training and validation cohorts. After that, multivariate Cox regression analysis was performed to adjust for some confounding factors. The results reveled that the risk score remained an independent predictor of OS for BC patients.
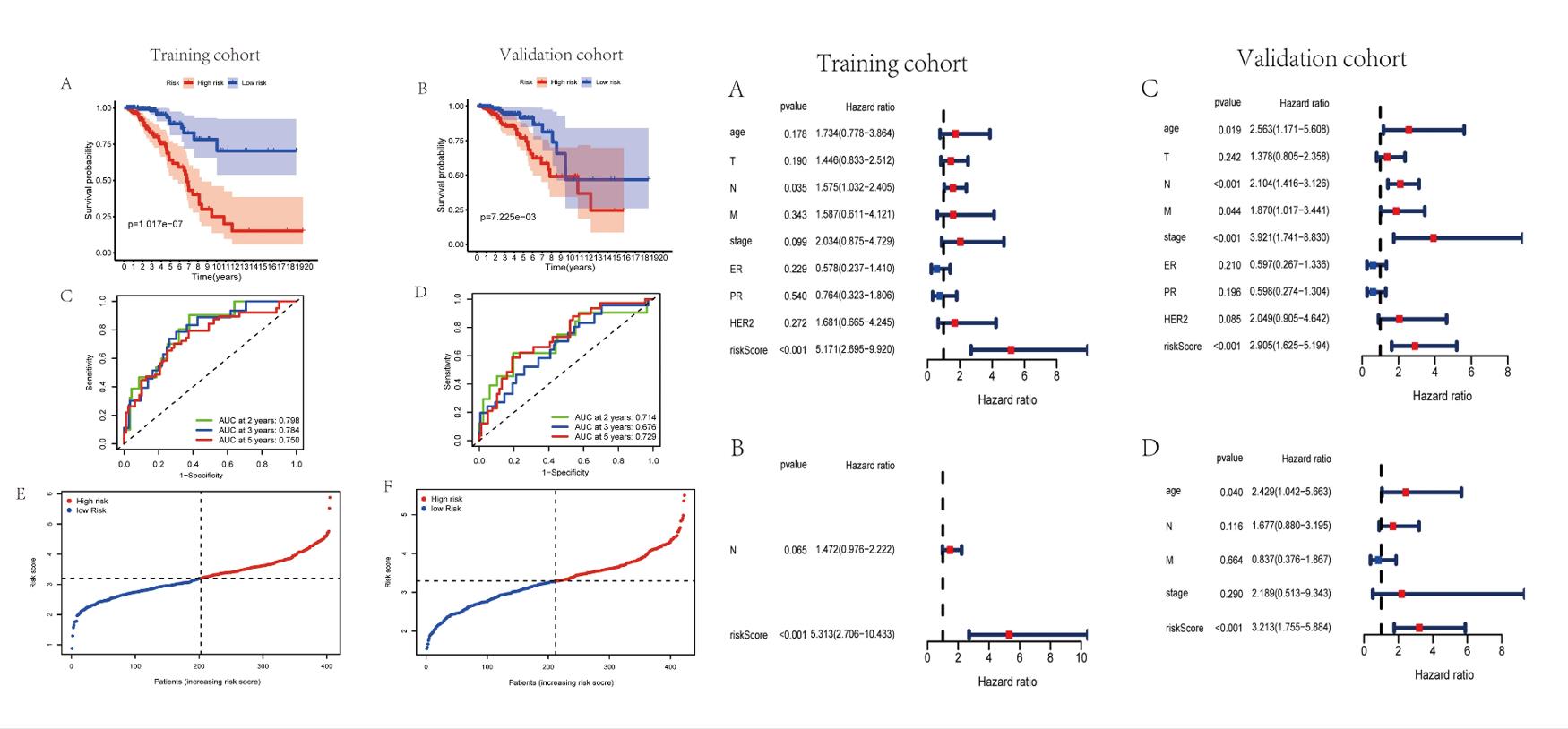 Fig. 2 Left: Evaluation of exosome-associated lncRNA risk model in training set and validation set; Right: Assessment of exosome-associated lncRNAs risk score as the independent prognostic factor in BC.
Fig. 2 Left: Evaluation of exosome-associated lncRNA risk model in training set and validation set; Right: Assessment of exosome-associated lncRNAs risk score as the independent prognostic factor in BC.
- The Sankey diagram not only presented the association between 15 exosome-associated lncRNAs and targeted mRNAs, but also presented the correlation between exosome-associated lncRNAs and the risk types consisted of risk or protective factors. Majorities of GO terms and KEGG pathways were identified associated with immune activation and response. Patients in high-risk group were demonstrated with lower stromal scores, immune scores and ESTIMATE scores in training and validation cohorts.
- The transcriptional expression of obvious mismatch repair genes in tumor tissues were calculated, resulting that MSH2, MSH6, MLH1, and PMS2 were all expressed markedly lower in the low-risk set. Eventually, the TCIA database was applied to calculate the IPS for each sample. The IPS of anti-CTLA-4, anti-PD-1, and anti-CTLA-4 plus anti-PD-1 were higher in the low-risk group, which strongly forecasted that patients in low-risk group had better immunotherapy responses.
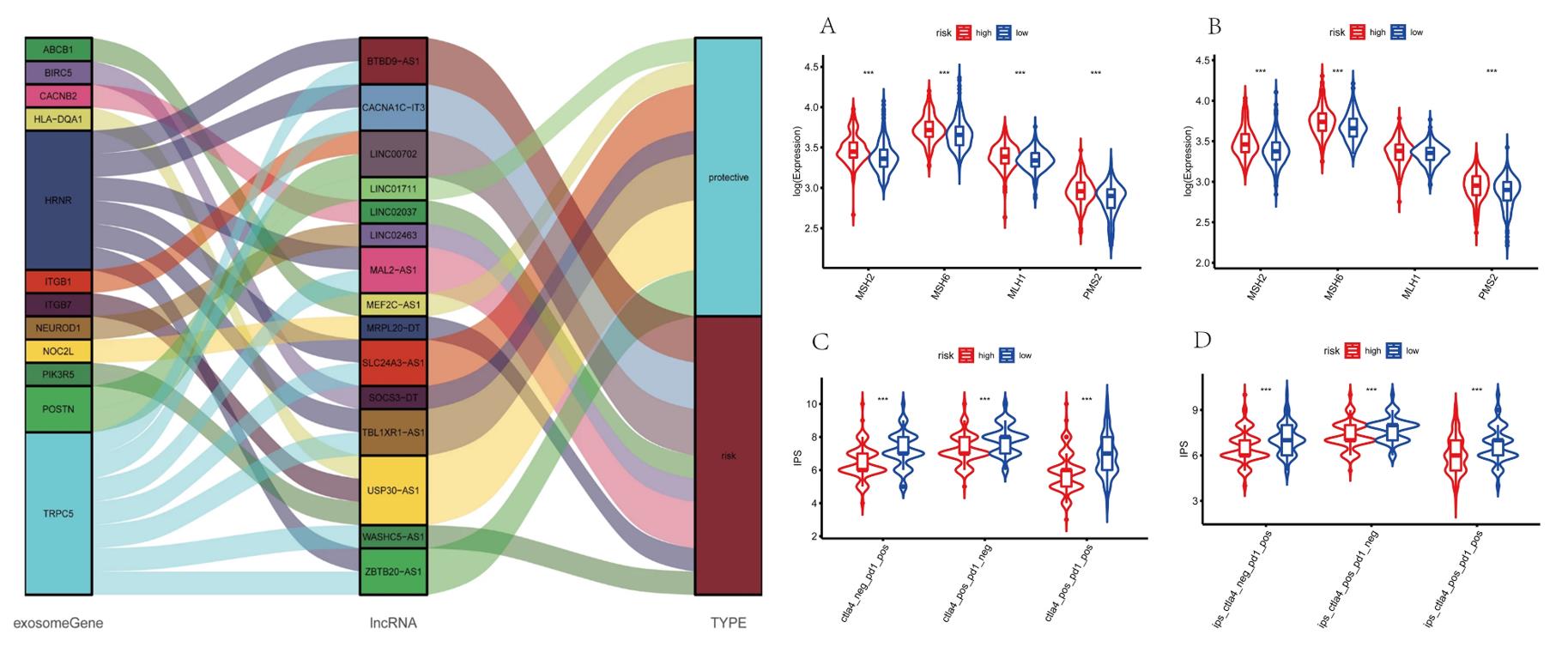 Fig. 3 Left: Construction of a LncRNA-mRNA co-expression network; Right: Prediction of immunotherapy response.
Fig. 3 Left: Construction of a LncRNA-mRNA co-expression network; Right: Prediction of immunotherapy response.
SUMMARY
- A novel exosome-related lncRNA risk model was established through bioinformatics approaches and relevant algorithms, that was related to the immune cell infiltration.
- Our study indicated that the exosome-related lncRNA-based signature showed outstanding performance in determining the prognosis and immunotherapy responsiveness of BC patients. Moreover, it can be served as a potential independent prognostic factor and provide novel insights for immunotherapy for BC.
RELATED PRODUCTS & SERVICES
Reference
- Qiu P, et al. (2022). "An exosome-related long non-coding RNAs risk model could predict survival outcomes in patients with breast cancer." Sci Rep. 12 (1), 22322.