Multi-Omics Approaches to Explore Immune Cell Functions
iScience. 2023 Mar 09; 26 (4): 106359.
Authors: Wang X, Fan D, Yang YQ, Gimple RC, Zhou ST.
INTRODUCTION
- When attacked by pathogens, the immune system orchestrates comprehensive responses including activation of and communication between a diverse series of immune cells. The characterization of each process and their associated products is indispensable for exploring the functions of immune cells and unraveling the underlying mechanisms in immune system responses.
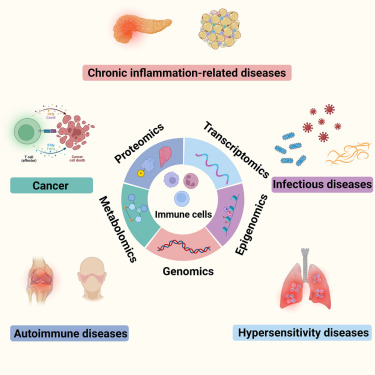 Fig. 1 Graphical abstract. (Xu W, et al., 2023)
Fig. 1 Graphical abstract. (Xu W, et al., 2023)
- As modern biological sciences evolve from investigation of individual molecules and pathways to growing emphasis on global and systems-based processes, increasing efforts have focused on combining the study of genomics with that of the other omics technologies, including epigenomics, transcriptomics, quantitative proteomics, global analyses of post-translational modifications (PTMs) and metabolomics, to characterize specific biological or pathological processes.
Genomics
- Whole genome sequencing (WGS) is a technique which aims to obtain all genes of a person including both coding and non-coding regions of a genome.
- Immuno-chip is a genotyping chip technique especially used for identifying significant loci for variants in immune-related diseases.
- Single-cell DNA sequencing (scDNA-seq) is a genomic approach used to decode variants, DNA modifications, and structural features of DNA at single cell level which may be ignored by using bulk DNA sequencing.
Epigenomics
- Chromatin immunoprecipitation followed by sequencing (ChIP–seq) is applied to profile the genome-wide features of DNA modifications including DNA methylation, histone modifications such as acetylation and methylation among others.
- DNase I hypersensitive sites sequencing (DNase-seq) is used to map the genome-wide active regulatory regions which are sensitive to DNase I.
- 3D genome technologies aim to study the three-dimensional (3D) structure of DNA and gene regulation, including chromosome conformation capture (3C), circularized chromosome conformation capture (4C), carbon copy chromosome conformation capture (5C), and Hi-C technology.
Transcriptomics
- Bulk RNA-sequencing, which is often referred as RNA-seq, is the most commonly used transcriptomic approach in which all RNAs from mixed samples including tissue, organ or a population of cells are extracted and sequenced through high-throughput technologies. The data from bulk RNA-seq represent an average expression of genes.
- Single-cell RNA sequencing (scRNA-seq) is a transcriptomic approach in which all RNAs from single cells are extracted and sequenced through high-throughput technologies. In contrast with bulk RNA-seq, scRNA-seq focuses on a single cell level, which can be used for identifying new subtypes of cells and exploring the intracellular and extracellular interactions.
Spatial Transcriptomics
- In situ hybridization is a technique using a nucleotide probe labeled with radio material, fluorescence or other substance which can be shown visually to detect nucleotide sequences in cells, tissue section or bulk tissue.
- In situ sequencing is a technique in which mRNA is sequenced directly in fixed tissue section or cells by using fluorescent probes.
- In situ capturing is a technique used to present the transcriptomics detain situ, which requires three steps to accomplish. Firstly, transcripts are captured and barcoded in the targeted tissue. Then, sequencing is completed outside the tissue. Finally, transcriptomics data is overlayed with the tissue image.
Proteomics and Post-Translational Modification
- Immunoassay (IA) is a commonly used method basing on antibody-antigen combination, which is applied in identifying and measuring molecules. Immunoassay is divided into non-labeled immunoassay (including western blot and immunodiffusion) and labeled immunoassay (including radioimmunoassay (RIA), enzyme-linked immunosorbent assay (ELISA), immunohistochemical methods, and fluorescence-activated cell sorting.
- Mass spectrometry (MS) is an analytical detection technique used to ascertain specific chemical substances including oligonucleotides, proteins, carbohydrates and other metabolites of cells by sorting and testing gaseous ions in electric and magnetic fields according to the mass-to-charge ratios.
- Cytometry time of flight (CyTOF), also called mass cytometry, is developed from flow cytometry which uses antibodies conjugated with heavy metal ion instead of fluorochromes to detect intracellular or extracellular antigens of interest on single cells.
- Single cell T cell receptor sequencing (scTCR-seq) is a method used to profile diversity and clonality of TCR repertoire and TCRαβ at single T cell level by multiplex PCR and high-throughput sequencing or Sanger sequencing.
RELATED PRODUCTS & SERVICES
Reference
- Wang X, et al. (2023). "Integrative Multi-Omics Approaches to Explore Immune Cell Functions: Challenges and Opportunities." IScience. 26 (4), 106359.