Microfluidic Aptamer-Based Devices for CTC Capture
Currently, aptamer-based technologies are widely studied in various biomedical fields. Over 6000 published articles on aptamers are referenced in the PubMed database, and more than 1000 of them are related to cancer. The first microfluidic device utilizing the aptamer to EGFR for isolating cancer cells from a laminar flow of human mononuclear cells has been reported by Wan et al. Two following microfluidic devices will be introduced.
The first device utilized elliptical (30 μm × 15 μm) micropillars 32 μm in height and an interpillar distance of 80 μm in flow channels (Fig.1). The micropillars were coated with DNA aptamers previously selected against the following cancer cell lines: CCRF- CEM cells, Ramos cells, HCT 116 cells, and DLD-1 cells. Less than 30 min was enough to isolate as few as 10 colorectal tumor cells from 1 ml of unprocessed whole blood using this small device. The cancer cells captured with the micropillar flow chamber were viable and used for following cellular and molecular characterization.
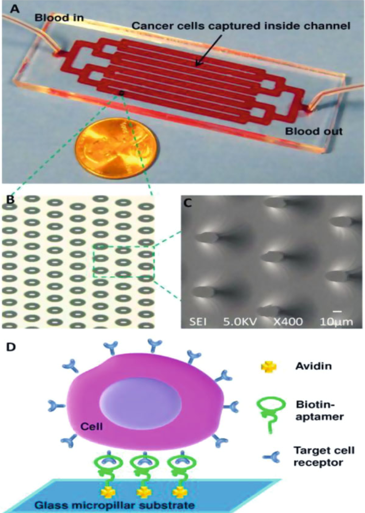 Figure 1. Microfluidic micropillar device for cancer cell isolation (a) Optical micrograph (b) and scanning electron microscope images of a micropillar array in a channel in the glass substrate (c) Scanning electron microscopy image of elliptical micropillars (d) Scheme of capturing cancer cells in the device. (Sheng et al. 2012)
Figure 1. Microfluidic micropillar device for cancer cell isolation (a) Optical micrograph (b) and scanning electron microscope images of a micropillar array in a channel in the glass substrate (c) Scanning electron microscopy image of elliptical micropillars (d) Scheme of capturing cancer cells in the device. (Sheng et al. 2012)
The second Hele-Shaw microfluidic device with plain surface of the channel and with array of pits on the channel floor have been utilized for the efficient detection of rare cancerous cells (Fig. 2). The pits were filled with anti-EGFR aptamer functionalized glass beads. Cancer cells, flowing in solution through the channel, were captured by the beads with high specificity. Such design helped sorting cell sub-populations with varying EGFR expression. Cells were released from the beads by a complementary to oligonucleotide sequence. This approach isolated viable cells for further analysis.
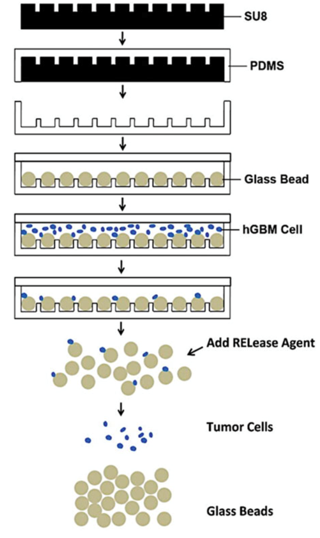 Figure 2. Scheme of fabrication and application of Hele-Shaw microfluidic device. SU-8 photoresist is spin-cast on silicon wafer, exposed and wells are developed to form the desired pattern. PDMS is poured on SU-8 master, baked, and peeled off. 50 μm diameter glass beads (GBs) are loaded into 25 μm deep pits and the substrate is covered with a flat PDMS slab. Cancer cell suspension is flowed through the device, and cells are captured by aptamers- functionalized GBs. Captured cells are finally released from the GB surface after GBs are collected from the device. (Wan et al. 2012)
Figure 2. Scheme of fabrication and application of Hele-Shaw microfluidic device. SU-8 photoresist is spin-cast on silicon wafer, exposed and wells are developed to form the desired pattern. PDMS is poured on SU-8 master, baked, and peeled off. 50 μm diameter glass beads (GBs) are loaded into 25 μm deep pits and the substrate is covered with a flat PDMS slab. Cancer cell suspension is flowed through the device, and cells are captured by aptamers- functionalized GBs. Captured cells are finally released from the GB surface after GBs are collected from the device. (Wan et al. 2012)
A flat channel microfluidic device made of polydimethylsiloxane and functionalized with locked nucleic acid (LNA) aptamers targeting EpCAM and nucleolin has been developed by Maremanda et al. for quick and efficient capture of CTCs and cancer cells. The analysis of blood samples obtained from 25 head and neck cancer patients detected as small as 5 ± 3 CTCs in ml of blood. These microfluidic devices also maintained viability for in vitro culture and characterization.
Recently, another microfluidic assay has been developed using a cocktail of aptamers with a synergistic effect. When a single aptamer was employed in the chip composed of silicon nanowires and an overlaid PDMS chaotic mixer, the capture affinity of the device was relatively weak. Nevertheless, using several aptamers, the synergistic effects among individual aptamers lead to an enhanced capture affinity (Fig.3). It has been shown that for the patients with nonsmall cell lung cancer (NSCLC) this method provided more comprehensive information in treatment monitoring.
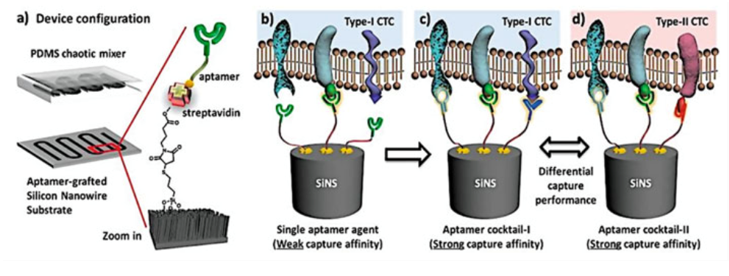 Figure 3. Scheme of an aptamer cocktail based CTC assay. Microfluidic CTC chip is composed of an aptamer-grafted silicon nanowire substrate and an overlaid PDMS chaotic mixer (a) When a single aptamer capture agent was employed, the capture affinity of the device is relatively weak for the lack of synergistic binding (b) By using cocktail capture agents, the synergistic effects among individual aptamers lead to an enhanced capture affinity (c) Different cocktail capture agents are expected to have differential capture performance for CTC subpopulation recognition. (Zhao et al. 2016)
Figure 3. Scheme of an aptamer cocktail based CTC assay. Microfluidic CTC chip is composed of an aptamer-grafted silicon nanowire substrate and an overlaid PDMS chaotic mixer (a) When a single aptamer capture agent was employed, the capture affinity of the device is relatively weak for the lack of synergistic binding (b) By using cocktail capture agents, the synergistic effects among individual aptamers lead to an enhanced capture affinity (c) Different cocktail capture agents are expected to have differential capture performance for CTC subpopulation recognition. (Zhao et al. 2016)
References
- Wan Y.; et al. Velocity effect on aptamer-based circulating tumor cell isolation in microfluidic devices. J Phys Chem B.2011, 115(47):13891–13896.
- Sheng W.; et al. Aptamer-enabled efficient isolation of cancer cells from whole blood using a microfluidic device. Anal Chem. 2012, 84(9):4199–4206.
- Martin J.; et al. Capturing cancer cells using aptamer-immobilized square capillary channels. Mol BioSyst. 2011, 7(5):1720–1727.
- Sefah K.; et al. DNA aptamers as molecular probes for colorectal cancer study. PLoS One. 2010, 5(12):e14269.
- Wan Y.; et al. Capture, isolation and release of cancer cells with aptamer- functionalized glass bead array. Lab Chip. 2012, 12(22):4693–4701.
- Maremanda N.; et al. Quick chip assay using locked nucleic acid modified epithelial cell adhesion molecule and nucleolin aptamers for the capture of circulating tumor cells. Biomicrofluidics. 2015, 9(5):054110.
- Zhao L.; et al. Enhanced and differential capture of circulating tumor cells from lung cancer patients by microfluidic assays using aptamer cocktail. Small. 2017,12 (8):1072–1081.