GUIDELINE
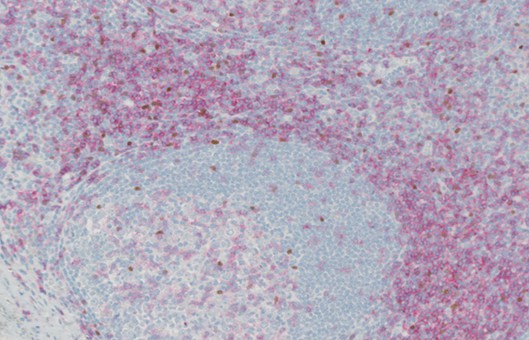
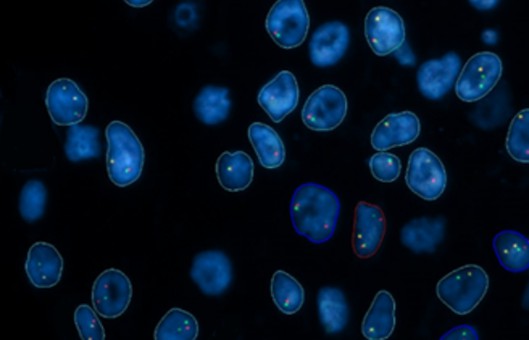
- In situ hybridization (ISH) is a type of hybridization that uses a labeled complementary DNA or RNA strand (i.e., probe) to localize a specific DNA or RNA sequence in a portion or section of tissue (In Situ) or in the entire tissue (whole mount ISH). Localization of endogenous transcripts is a desirable approach for confirming expression patterns.
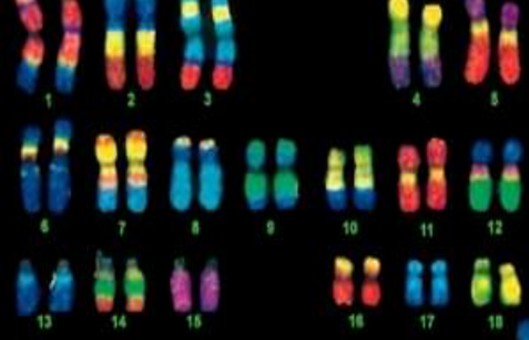
- DNA ISH can be used to determine the structure of chromosomes. However, RNA ISH (hybridization histochemistry) is used to measure and localize mRNAs and other transcripts within tissue sections or whole mounts.
METHODS
- Deparaffinization. For frozen sections, start at Step 2. If using formaldehyde-fixed paraffin-embedded sections, continue with this step.
- Antigen retrieval. Digest with 20 µg/mL proteinase K in pre-warmed 50 mM Tris for 10-20 min at 37°C. Incubation time and proteinase K concentration may require optimization. Rinse slides 5x in distilled water.
- Immerse slides in ice-cold 20% (v/v) acetic acid for 20 sec. This permeabilizes the cells to allow access to the probe and the antibody.
- Dehydrate the slides by washing for approximately 1 min per wash in 70% ethanol, 95% ethanol and 100% ethanol, then air dry.
- Add 100 µL of hybridization solution to each slide.
- Incubate the slides for 1 h in a humidified hybridization chamber at the desired hybridization temperature. Typical hybridization temperatures range between 55-62°C.
- Dilute the probes in hybridization solution in PCR tubes. Heat at 95°C for 2 min in a PCR block to denature the RNA or DNA probe. Chill on ice immediately to prevent reannealing.
- Drain off the hybridization solution. Add 50–100 μL of diluted probe per section, covering the entire sample. Incubate in the humidified hybridization chamber at 65°C overnight. Cover the sample with a cover slip to prevent evaporation.
- Wash twice in MABT (maleic acid buffer containing Tween 20) for 30 min at room temperature. MABT is gentler than PBS and is more suitable for nucleic acid detection. Dry the slides.
- Transfer to a humidified chamber and add 200 µL blocking buffer to each section (MABT + 2% BSA, milk or serum). Block for 1-2 h at room temperature.
- Drain off the blocking buffer. Add the anti-label antibody at the required dilution in blocking buffer. Check the antibody datasheet for a recommended concentration/dilution. Incubate for 1-2 h at room temperature.
- Wash slides 5x10 min with MABT at room temperature.
- Wash the slides 2x10 min each at room temperature with pre-staining buffer (100 mM Tris pH 9.5, 100 mM NaCl, 10 mM MgCl2).
- For fluorescence detection, proceed to the next step. For other forms of detection, return the slides to the humidified chamber and follow manufacturer's instructions for color development. Rinse slides in distilled water.
- Air dry the slides for 30 min. Wash in 100% ethanol and air dry thoroughly.
- Mount using mounting solution.
NOTES
- The RNA probe will hybridize to the corresponding mRNA, or the DNA probe will hybridize to the corresponding cellular DNA. Optimize the hybridization temperature depending on the sequence of the probe used, as well as the cell or tissue type. This temperature should be optimized for each tissue type analyzed.
- For very short DNA/RNA probes (0.5-3 kb) or very complex probes, the washing temperature should be lower (up to 45°C) and the stringency lower (1-2x SSC). For single-locus or large probes, the temperature should be around 65°C and the stringency high (below 0.5x SSC). The temperature and stringency should be highest for repetitive probes (such as alpha-satellite repeats).
RELATED PRODUCTS & SERVICES