A Reference Human Induced Pluripotent Stem Cell Line
Cell Stem Cell. 2022 Dec 1; 29(12): 1685-1702
Authors: Pantazis CB, Yang A, Lara E, McDonough JA, Blauwendraat C, Peng L, Oguro H, Kanaujiya J, Zou J, Sebesta D, Pratt G, Cross E, Blockwick J, Buxton P, Kinner-Bibeau L, Medura C, Tompkins C, Hughes S, Santiana M, Faghri F, Nalls MA, Vitale D, Ballard S, et al.
INTRODUCTION
Human induced pluripotent stem cells (iPSCs) are increasingly used to model diseases because they capture genetic contributors to disease risk and can be differentiated into relevant cell populations. Additionally, the genomes of iPSCs can be edited to introduce or correct disease-associated variants. However, the considerable phenotypic variation between lines makes it challenging to replicate key findings and integrate data across research groups.
METHODS
- The study focused on a subset of lines with broad consents for data sharing and further dissemination of the line and its derivatives, and identified KOLF2_C1, KUCG3, LNGPI1, NCRM1, NCRM5, NN0003932, NN0004297, and PGP1. We selected one to four sub-lines per parental cell line for further expansion based on their typical stem cell morphology under phase contrast microscopy and normal karyotype from the analysis of >20 Giemsa-band metaphase chromosome spreads. We then selected a single clonal sub-line from each parental line for further expansion into 192 replicate stock vials.
- To understand the genetic background of each sub-line, the study sequenced their genomes at >30× coverage. They examined WGS data for known pathogenic mutations in ADRD-associated genes, and tested for the presence of genetic variants that have a strong to moderate association with ADRD. Finally, they calculated polygenic risk scores (PRSs) for all iPSC lines based on their cumulative burden of common genetic variants associated with AD or PD.
- We next characterized the efficiency with which an SNV could be introduced. Using improved conditions for homology-directed repair, we introduced a G in place of the wild-type (WT) C in exon 1 of the TIMP3 gene located at Chr22q12.3, resulting in an S38C missense mutation. Twenty-four edited clones from each sub-line were then genotyped by Sanger sequencing of PCR amplicons spanning the targeted site of the TIMP3 locus.
- To rigorously benchmark the performance of KOLF2.1J relative to other lines, we distributed it to other groups, nine of whom returned quantitative information on the performance of KOLF2.1J in head-to-head comparisons with 12 other iPSC lines across 10 differentiation protocols or assays established in their laboratories.
RESULTS
- Each sub-line had the morphology expected for human iPSCs, including a high nuclear to cytoplasmic ratio, prominent nucleoli, growth in colonies with well-defined borders, and an absence of differentiated cells, but they are varied in their survival and proliferation rates. All analyzed iPSC sub-lines had gene expression patterns consistent with iPSCs, and six of the seven genetically distinct sub-lines showed similar transcriptional profiles.
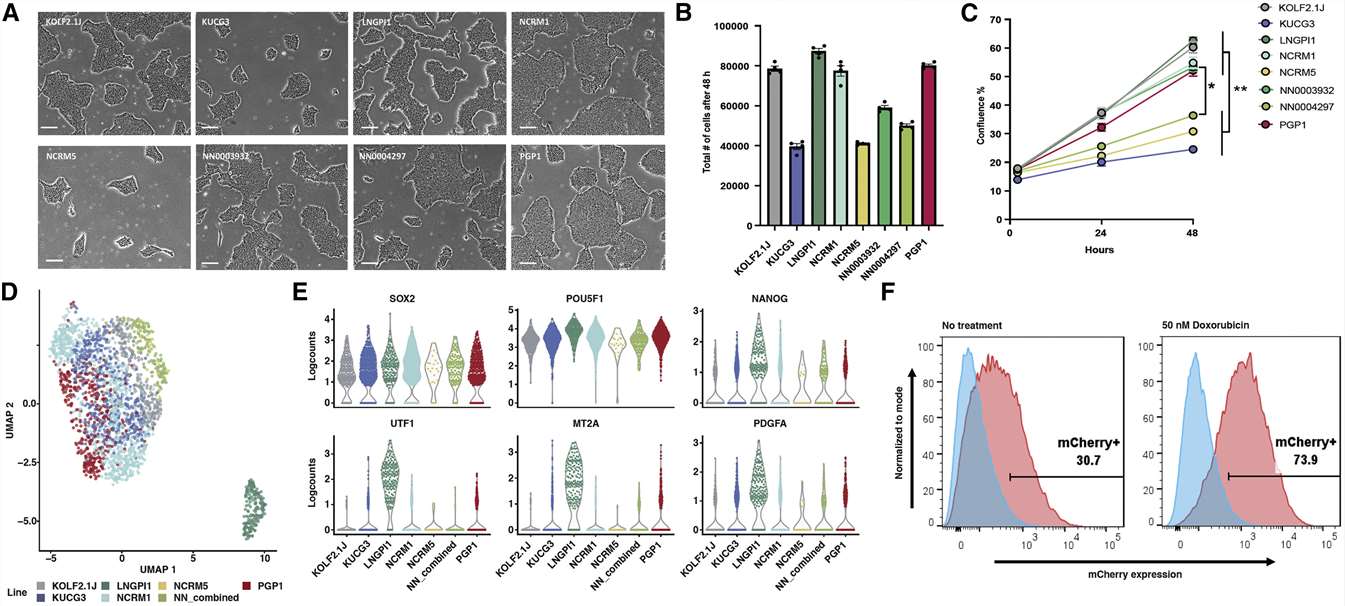 Fig. 1 Growth, gene expression, and p53 pathway integrity of candidate cell sub-lines.
Fig. 1 Growth, gene expression, and p53 pathway integrity of candidate cell sub-lines.
- The distribution of insertion-deletion (indel), loss-of-function (LoF), and missense SNVs was similar across all lines and similar to human populations in the gnomAD database. A modest number of such potentially deleterious variants per cell line, and the PRSs of all iPSC lines fall within the expected range of the population.
- We quantify frequency of six possible genotypes, WT/WT, WT/SNV, WT/indel, SNV/SNV, SNV/indel, and indel/indel. Homozygous (SNV/SNV) genotypes were produced at an overall efficiency of over 40%. We also found very high concordance across the genome when comparing gene-edited clones with their parental sub-lines using DNA microarray data, suggesting that the number of SNVs acquired during CRISPR editing was lower than the theoretical limit set by the call rates for this array. Except for KUCG3, tested iPSC sub-lines do not contain mosaic populations of large genetic variants or readily acquire them upon gene editing under the conditions we tested.
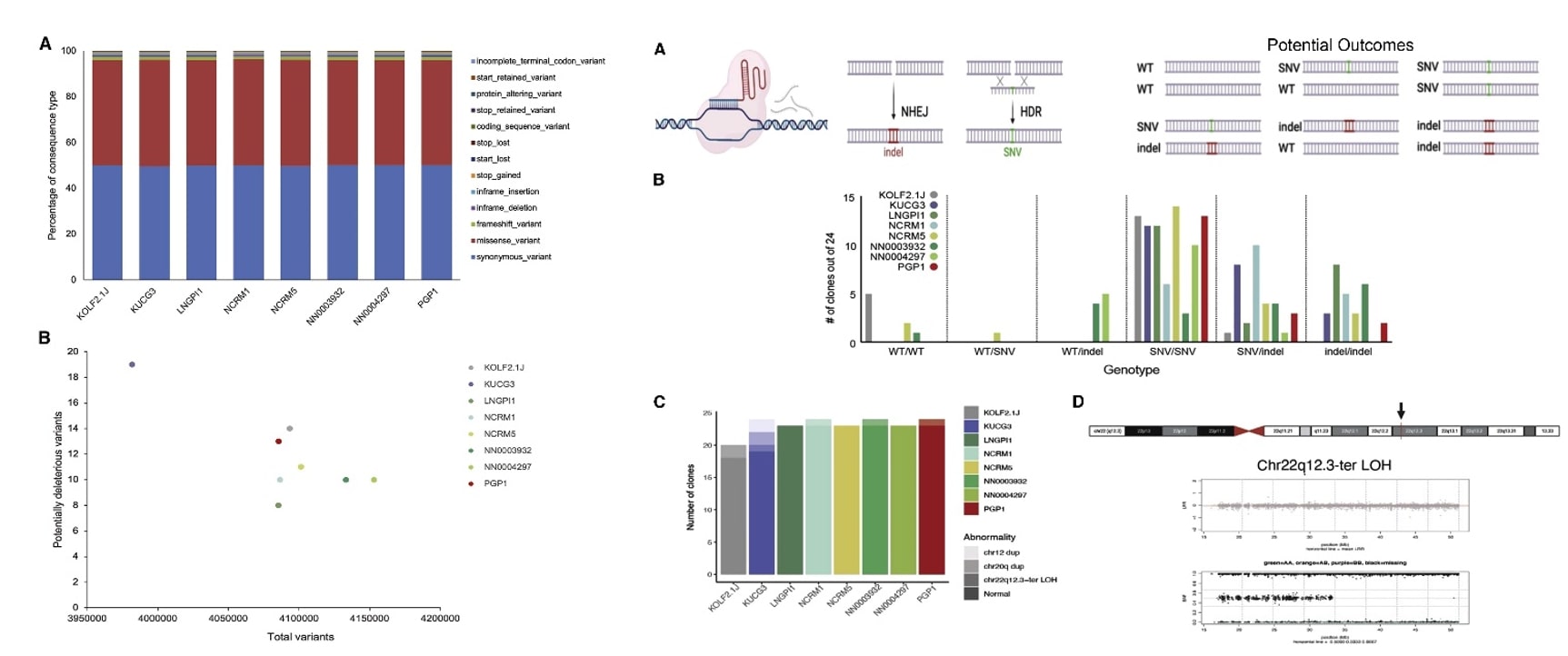 Fig. 2 Left: Genetic analyses of 8 candidate iPSC sub-lines; Right: Comparative gene editing efficiency.
Fig. 2 Left: Genetic analyses of 8 candidate iPSC sub-lines; Right: Comparative gene editing efficiency.
- Because KOLF2.1J performed well across all tested assays and was found to harbor APOE3/E3, we selected it as a candidate reference iPSC sub-line. KOLF2.1J was differentiated into cells from all three germ layers, cortical glutamatergic neurons, cortical forebrain neurons, skeletal myocytes, motor neurons, astrocytes, microglia and macrophages,and dopaminergic progenitors, neurons, and organoids, using established differentiation protocols.
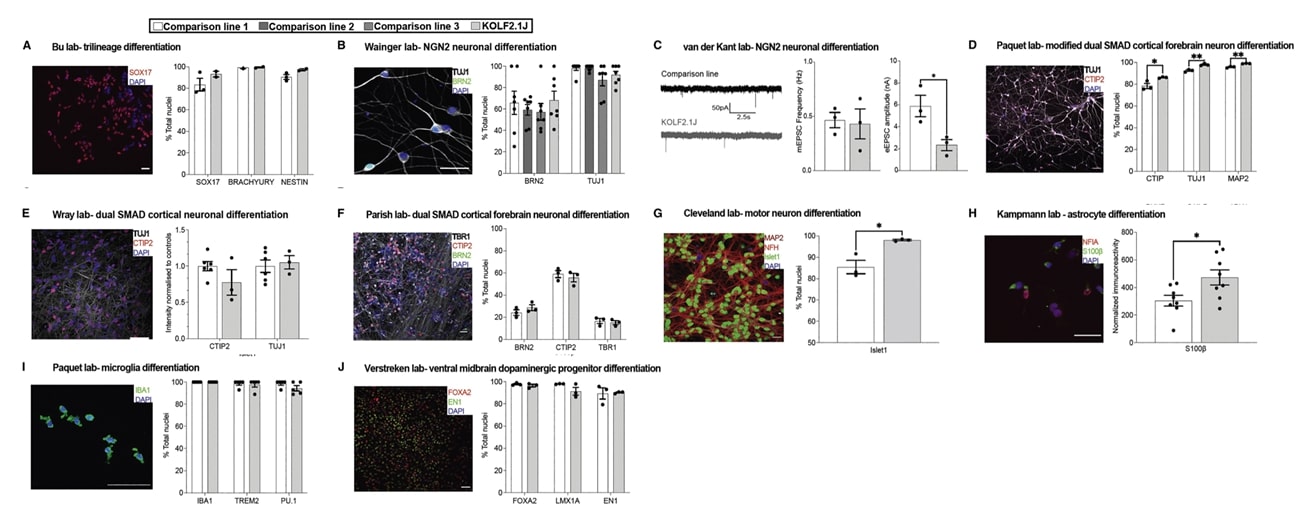 Fig. 3 Performance of KOLF2.1J and established hPSC lines in head-to-head comparisons.
Fig. 3 Performance of KOLF2.1J and established hPSC lines in head-to-head comparisons.
SUMMARY
- The study sub-cloned candidate human iPSC lines and deeply characterized their genetic properties using whole genome sequencing, their genomic stability upon CRISPR-Cas9-based gene editing, and their phenotypic properties including differentiation to commonly used cell types.
- The study identified KOLF2.1J as an all-around well-performing iPSC line. On the strength of various findings, they have made KOLF2.1J and its gene-edited derivative clones readily accessible to promote the standardization required for large-scale collaborative science in the stem cell field.
RELATED PRODUCTS & SERVICES
Reference
- Pantazis CB, et al. (2022). "A reference human induced pluripotent stem cell line for large-scale collaborative studies." Cell Stem Cell. 29 (12), 1685-1702.