Screening for NRG1 Gene Rearrangements by FISH
Diagnostic Pathology. 2024 Jan 3; 19 (1): 1.
Authors: Zhang X, Li L, Gao F, Liu B, Li J, Ren S, Peng S, Qiu W, Pu X, Ye Q.
INTRODUCTION
NRG1 is a member of the neuregulin (NRG) complex family, which is composed of six structurally related cellular growth factors encoded by six closely related genes (NRG1-NRG6). Although NRG1 gene rearrangement is rare (~ 0.1-0.3%), it has been reported as a potentially actionable genomic event observed in various tumor types. NRG1 fusions can promote pathological signaling via MAPK and other canonical pathways when present.
METHODS
- Each tissue sample underwent immediate fixation in 10% neutral buffered formalin for a duration of 12-48 h, followed by paraffin embedding. The processed samples were subjected to routine deparaffinization and rehydration procedures. A tissue microarray (TMA) was created using an automated arrayer with a 2 mm punch size obtained from representative tumor blocks of each case.
- Four-micrometer-thick, formalin-fixed, and paraffin-embedded tissue sections were used for FISH. FISH testing for NRG1 gene rearrangements was performed using the NRG1 probes. FISH positivity was defined when more than 15% of tumor cells displayed signals with distinct red and green signals or a signal pattern that maintained a single red signal.
- IHC staining was carried out on an automatic system using an automated staining protocol. IHC staining scores were calculated by multiplying the staining intensity by the percentage of immunoreactive tumor cells (0 to 100).
- Browse our recommendations
| Product/Service Types | Description |
| Tissue Arrays | Creative Bioarray provides a comprehensive range of pre-made and custom tissue arrays. |
| Fluorescent In Situ Hybridization (FISH) Service | Creative Bioarray offers a range of different FISH services including metaphase and interphase FISH, fibre-FISH, RNA-FISH, M-FISH, 3D-FISH, flow-FISH, FISH on paraffin sections, and immune-FISH. |
| Probe | Creative Bioarray provides the most comprehensive list of FISH probes for rapid identification of a wide range of chromosomal aberrations across the genome. |
| Immunohistochemistry (IHC), Immunofluorescence (IF) Service | Creative Bioarray offers a comprehensive IHC service from project design, and marker selection to image completion and data analysis. We are dedicated to satisfying every customer and assisting them to achieve their specific research goals. |
RESULTS
- The NRG1 dual color break-apart probe was specifically designed to identify the occurrence of translocations within the chromosomal region 8p12 that contains the NRG1 gene. The green probe covered NRG1 exons 1-2 and the chromosomal region preceding the 5' end of the NRG1 gene, while the orange probe was responsible for identifying the chromosomal region following the 3' end of the NRG1 gene. Concerning FISH signals, three distinct types of abnormalities were observed: (A) break-apart signal (three cases) with or without high copy number of the 3'-end of the gene; (B) low copy number of the 5'-end of the gene concerning the 3'-end of the gene, with fusion signals (12 cases); and (C) low copy number of the 5'-end of the gene concerning the 3'-end of the gene, without fusion signals.
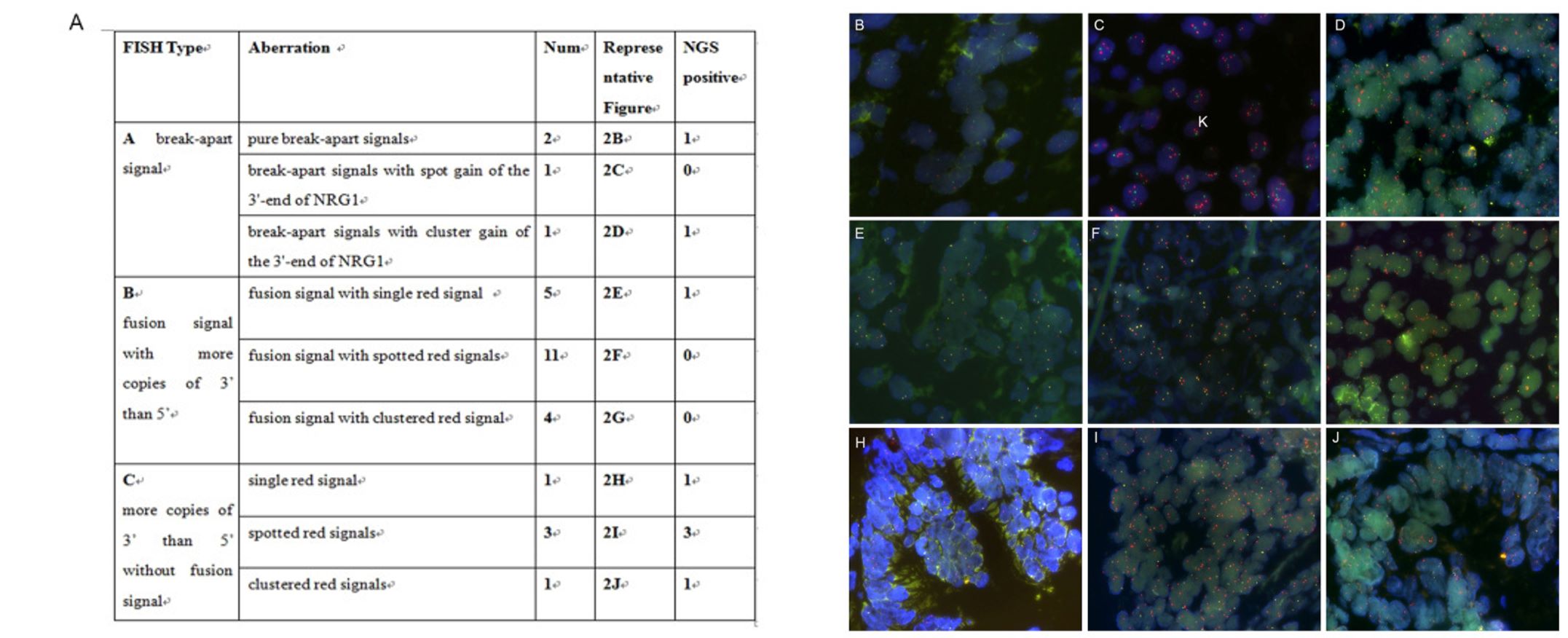 Fig. 1 Distinct FISH signal types of NRG1 aberrations.
Fig. 1 Distinct FISH signal types of NRG1 aberrations.
- To validate the FISH results, all of the FISH-positive cases underwent further testing using NGS (both DNA- and RNA-based). However, the results of the FISH analysis were not in agreement with the NGS results. Out of the 29 cases with abnormal NRG1 FISH signals, only eight cases were conclusively proven to have NRG1 fusion through the use of NGS. Break-apart signals or fusion signals possessing more copies of 3' than 5' NRG1 do not necessarily indicate the presence of true fusions.
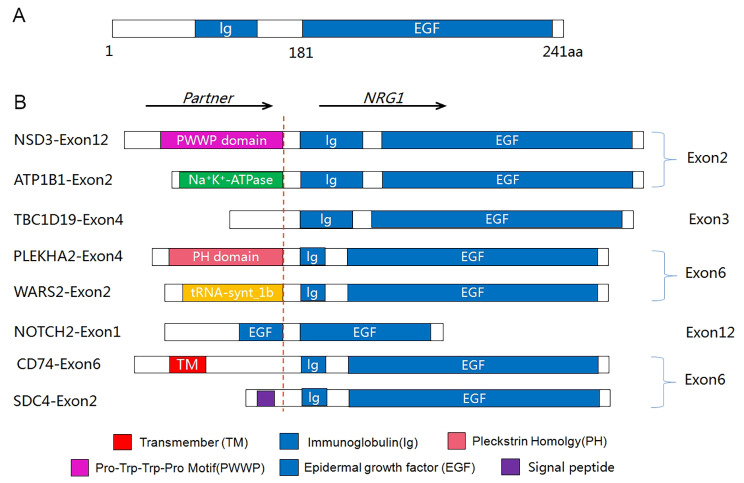 Fig. 2 Schematic diagram of NRG1 fusion variants in pan-cancers.
Fig. 2 Schematic diagram of NRG1 fusion variants in pan-cancers.
SUMMARY
Considering the high cost of NGS, FISH remains a useful method for screening NRG1 fusions in various types of tumors. This study provides valuable insights into the molecular mechanisms of NRG1 fusion and identifies potential treatment targets for patients suffering from this disease.
RELATED PRODUCTS & SERVICES
Reference
- Zhang X, et al. (2024). "Fluorescent in situ hybridization has limitations in screening NRG1 gene rearrangements." Diagn Pathol. 19 (1): 1.