In Vitro Cellular Reprogramming to Model Disorders of Sex Development
Science Advances. 2023 Jan 4; 9(1): eabn9793
Authors: Gonen N, Eozenou C, Mitter R, Elzaiat M, Stévant I, Aviram R, Bernardo AS, Chervova A, Wankanit S, Frachon E, Commère PH, Brailly-Tabard S, Valon L, Barrio Cano L, Levayer R, Mazen I, Gobaa S, Smith JC, McElreavey K, Lovell-Badge R, Bashamboo A.
INTRODUCTION
During embryonic development, mutually antagonistic signaling cascades determine gonadal fate toward a testicular or ovarian identity. Errors in this process result in disorders of sex development (DSDs), characterized by discordance between chromosomal, gonadal, and anatomical sex. The absence of an appropriate, accessible in vitro system is a major obstacle in understanding mechanisms of sex-determination / DSDs.
METHODS
- To enable differentiation of mouse embryonic stem cells (mESCs) toward gonadal progenitors, the study developed protocols that mimic the known pathway of gonad development, whereby pluripotent cells differentiate toward early mesoderm progenitors followed by further regional specification toward the mesoderm from which the gonads originate, which includes intermediate mesoderm (IM) and associated coelomic epithelium.
- To be able to compare bulk RNA-seq with the scRNA-seq data, the study generated pseudo bulks from the six identified cell clusters. They compared the gene expression of the bulk and the scRNA-seq data by performing pairwise Spearman correlations on all the protein-coding genes and on a set of marker genes of the six cell clusters. They then performed the comparison between the RNA-seq performed along the mESC reprogramming and the scRNA-seq data using both the protein-coding genes and the list of marker genes.
- The study next used three different hiPSC lines (two clones each), two from healthy individuals (46, XY male and 46, XX female) and one from a patient with 46, XY DSD that carries the pathogenic p. Arg313Cys variant in NR5A1. To further assess the ability of hiPSC-derived supporting cells to form tubular structures, they seeded the sorted cells atop a soft substrate made of Matrigel (50%, v/v). To evaluate the capacity of supporting cells derived from iPSCs to migrate, aggregate, and form 3D structures, they designed a polydimethylsiloxane (PDMS) microfluidic device termed GONAChip with three channels.
- The study corrected the pathogenic variant in 46, XY DSD hiPSCs using CRISPR-Cas9 genome editing. The edited cells were then subjected to differentiation following the same protocols as for the 46, XY and 46, XY DSD iPSCs.
RESULTS
- Cells were then differentiated to an epiblast stem cell (EpiSC)-like state by culture in N2B27 media for 2.5 days. Then, they were differentiated into early mesoderm progenitors by exposing the cells to a chemically defined media for 36 hours. Relying on the BMP4 gradient to which cells in the region of the IM cells are exposed, the study tested five different conditions (termed IM1 to IM5) and screened the cells by marker analysis, and identified IM3 is the optimal condition to differentiate the mESCs toward early gonadal progenitors.
- In vitro-derived gonadal progenitors can induce embryonic Sertoli cells (iSLCs). The study compares transcriptomes of in vitro-derived gonadal-like cells with cells isolated directly from in vivo gonads. The IM3 and IM4 show a strong correlation with early progenitor cells in both comparisons, and the iSLCs show a high correlation with both the early and interstitial progenitors, reflecting the similarity of their transcriptomes.
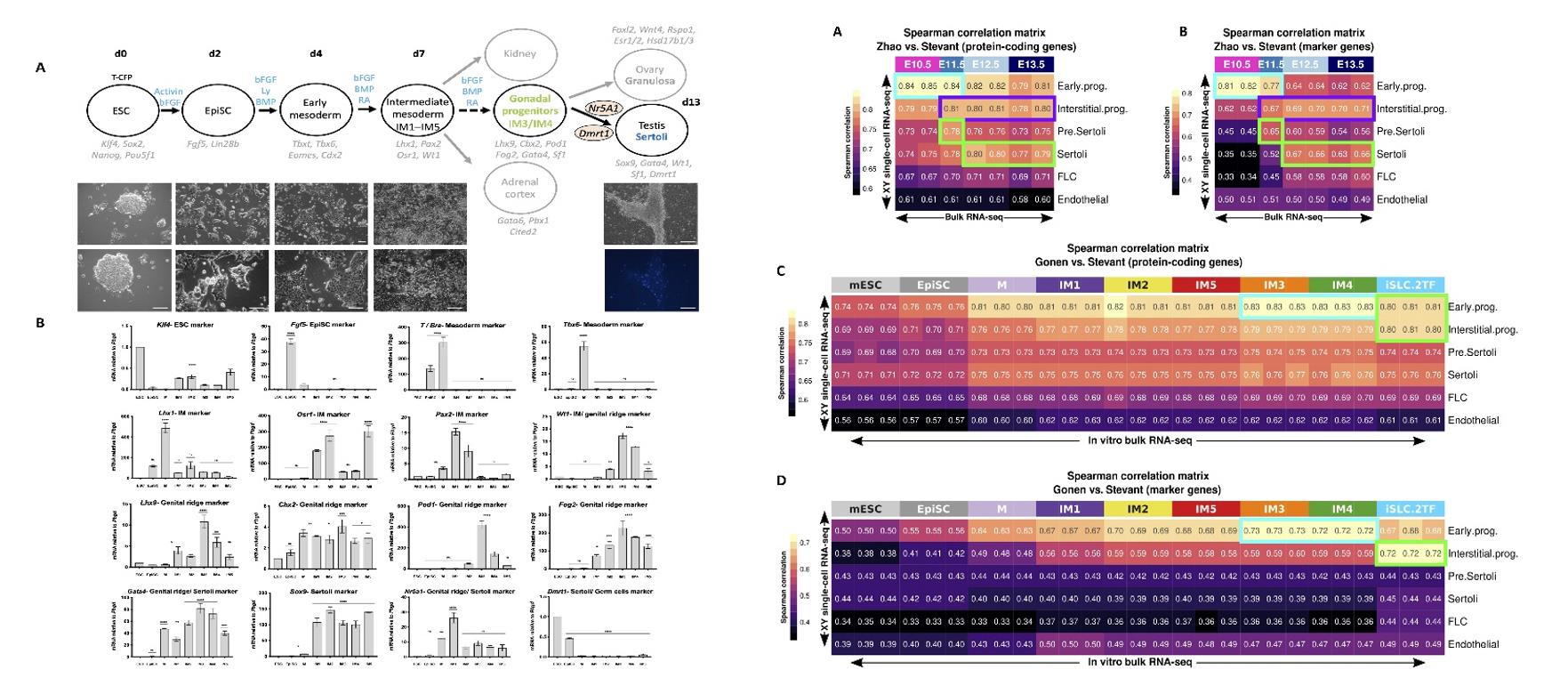 Fig. 1 Left: Differentiation of mouse ESCs toward gonadal cells; Right: Comparing the transcriptome of in vitro-derived gonadal cells with scRNA-seq of in vivo gonadal cells.
Fig. 1 Left: Differentiation of mouse ESCs toward gonadal cells; Right: Comparing the transcriptome of in vitro-derived gonadal cells with scRNA-seq of in vivo gonadal cells.
- The cells derived from 46, XY iPSCs can form tubular structures, whereas those derived from 46, XX and 46, XY-DSD hiPSCs aggregate but fail to form tubular structures. Besides, 46, XY and 46, XY DSD cells migrate and invade the central channel filled with Matrigel (hours 0 to 30), whereas the 46, XX cells do not. And 46, XY DSD invasion of Matrigel appears less coordinated (hours 30 to 60) and the cells do not form tubular structures.
- The 46, XY DSD cells after correction of the NR5A1 variant show spatial reorganization of CLAUDIN-11 and VIMENTIN similar to that of 46, XY cells. On prolonged culture (for over 7 weeks) the cells derived from this rescued cell line continue to express the SOX9 protein. The rescued cells show spontaneous tubule formation similar to those derived from 46, XY hiPSCs. Using the GONAChip device, sorted CLAUDIN-11-positive rescued cells proliferated, migrated (hours 0 to 30), and self-organized to form distinct 3D tubular structures (hours 30 to 60) similar to those derived from 46, XY DSD iPSCs.
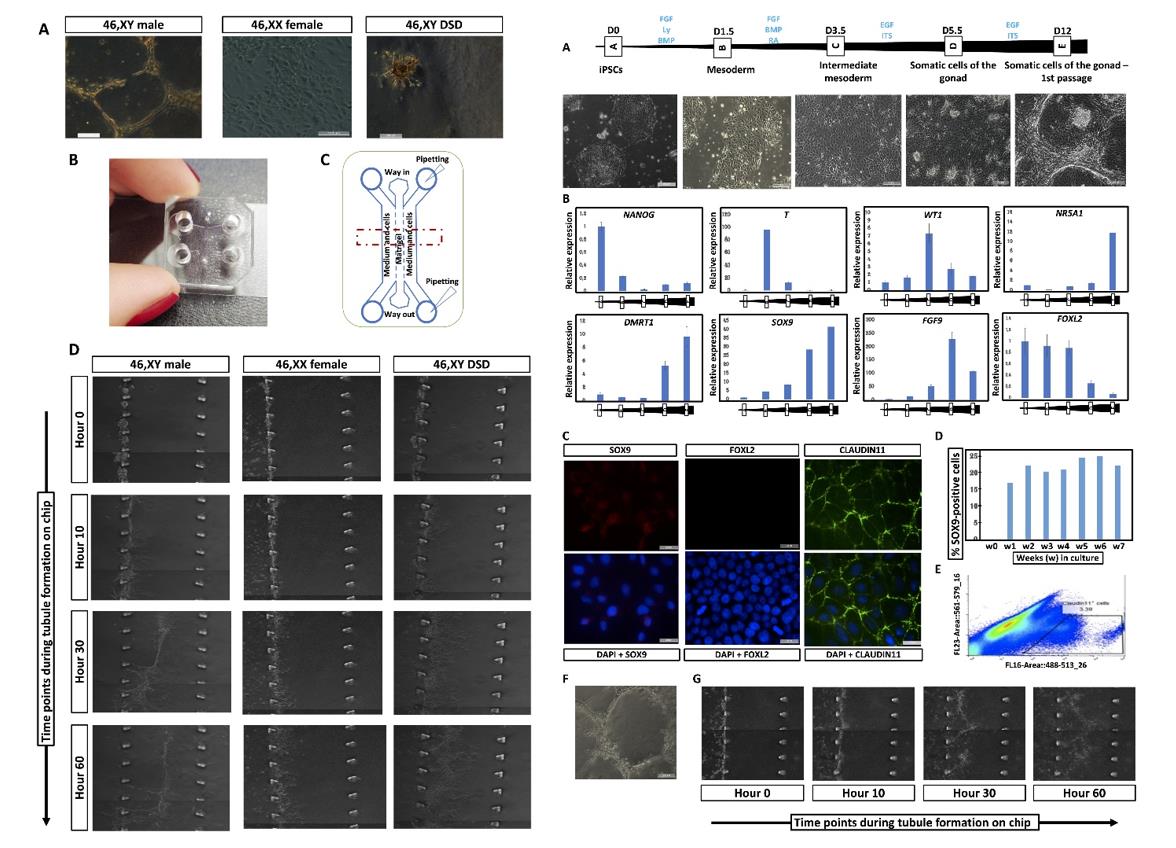 Fig. 2 Left: 3D tubule formation in a microfluidic chip. Right: CRISPR-CAS9 correction of the NR5A1 pathogenic variant in iPSC-derived 46, XY DSD cells restores iSLC properties.
Fig. 2 Left: 3D tubule formation in a microfluidic chip. Right: CRISPR-CAS9 correction of the NR5A1 pathogenic variant in iPSC-derived 46, XY DSD cells restores iSLC properties.
SUMMARY
- The study describes protocols for differentiation of mouse and human pluripotent cells toward gonadal progenitors. Transcriptomic analysis reveals that the in vitro-derived murine gonadal cells are equivalent to embryonic day 11.5 in vivo progenitors.
- Using similar conditions, Sertoli-like cells derived from 46, XY human induced pluripotent stem cells (hiPSCs) exhibit sustained expression of testis-specific genes, secrete anti-Müllerian hormone, migrate, and form tubular structures.
- Cells derived from 46, XY DSD female hiPSCs, carrying an NR5A1 variant, show aberrant gene expression and absence of tubule formation. CRISPR-Cas9-mediated variant correction rescued the phenotype. This is a robust tool to understand mechanisms of sex determination and model DSDs.
RELATED PRODUCTS & SERVICES
Reference
- Gonen N, et al. (2023). "In vitro cellular reprogramming to model gonad development and its disorders." Sci Adv. 9 (1), eabn9793.