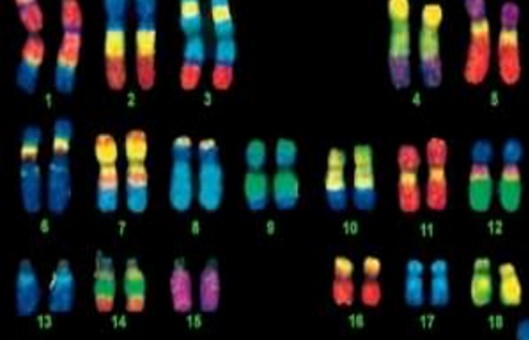
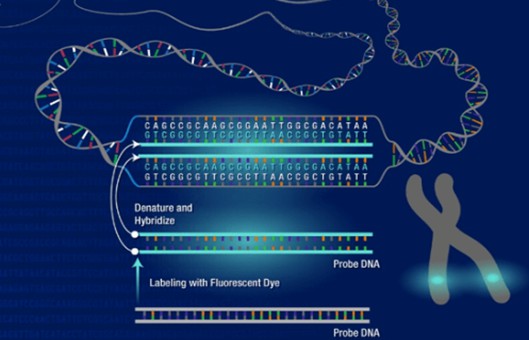
FISH Protocol for Transgene Mapping on G-Banded Mouse Chromosome
GUIDELINE
- The techniques for producing transgenic mice are now being used extensively, and it is often desirable to determine the site of transgene insertion. Although fluorescence in situ hybridization (FISH) permits the rapid and efficient localization of DNA probes on human chromosomes, it has not been effectively applied to the mapping of sequences on mouse chromosomes.
- A highly sensitive method for the mapping of transgenes and other genes in the mouse genome is described. This technique combines high-resolution G-banding and fluorescence in situ hybridization (FISH) with either biotin/avidin-FITC or digoxigenin-anti-digoxigenin-FITC, the latter being the more sensitive. Banding patterns are obtained with trypsin/Giemsa-treated slides, and sensitivity is greatly increased by the use of mouse Cot-1 DNA.
METHODS
Preparation of chromosomes
- Thirty-five to 70 μL of heparinized blood was obtained from the tail vein of 6- to 8-week-old mice, and cultures in a final volume of 1.5 ml lymphocyte culture media [RPMI 1640, 15% fetal bovine serum (heat inactivated at 56°C; 1h), 25 mM HEPES, 2.4 mM glutamine, 0.1 mg/ml, gentamycin sulfate, 6 μg/ml PHA HA16, and 3.3 μg/ml LPS] were incubated in a 37°C water bath.
- After 41-44 h, 5 μg of colchicine was added to each culture, and incubation was continued for another 10-20 min.
- The medium was changed when cultures were in log phase growth and nearly confluent (2-3 days after splitting).
- Velban was added to a final concentration of 5-10 ng/ml 24 h later, and incubation continued for an additional 3 h.
- The cultured cell suspension was transferred to 1.5 ml microcentrifuge tubes, the cells were pelleted for 5 min at 600 g in a variable speed microfuge, and the supernatant medium was removed.
- The cells were gently resuspended in 1.0 ml 0.56% KCI prewarmed to 37°C and incubated for 15 min in a 37°C water bath. Twenty microliters of fixative (3:1 methanol: acetic acid) were added to each tube, and the cells were again pelleted at 600 g.
- All but 50 μl of supernatant fluid was removed by aspiration, the cell pellets were gently resuspended in the remaining supernatant fluid, carefully overlaid with 1 ml fixative, gently mixed by inversion, and left for at least 30 min at room temperature.
- The fixed suspension of nuclei was further dehydrated by three cycles of fixation (pellet at 600 g, supernatant fluid aspirated, 0.5 ml fresh fixative added). After the third fixation, nuclei were resuspended in 100-200 μl fixative, dropped from a vertical height of 20 cm onto prewashed, water-flooded slides (25×75 mm slides rinsed in methanol and soaked in water), and allowed to air dry. Slides were stored at -70°C until used for banding and in situ hybridization.
Preparation of chromosomes
- Old (stored at -70°C for up to 2 months) or newly made slides were incubated at 2×SSC at 60°C for 1.5 h.
- The slides were then rinsed in three changes of 0.9% NaCl solution at room temperature for 5 min each with shaking and then stained for 6-8 min in a trypsin-Giemsa solution.
- The slides were then transferred to fresh buffer solution (Gurr buffer/distilled water, 1:1) for 2 min, rinsed individually in three changes of buffer solution, and air dried.
- After photography, the immersion oil was removed by treating the slides with 1:1 xylol/ethanol for 1 min. The slides were then de-stained with two changes of 3:1 methanol/acetic acid, 5 min each, air dried, postfixed with 3.7% formaldehyde (in PBS) for 15 min, washed in two changes of PBS, 5 min each, and air dried.
Labeling of DNA probe
- The probe used for localization of the human CuZnSOD (SOD1) transgenes was the entire plasmid pGhSOD1-SVneo containing a 14.5 kb EcoRI-BamHI genomic fragment encompassing human SOD1.
Creative Bioarray Relevant Recommendations
- If you have problems with FISH probes, please feel free to browse our Custom Probes. Tell us the specific gene or the chromosome region of your interest, and the desired dye color, and we will make a custom probe for you. This exciting new service enables the production of personalized solutions for patients and research projects anywhere and for anyone around the world.
In situ hybridization
- Incubate with 20 μl 100 ug/ml RNase A in 2×SSC under a coverslip (20 X 40 mm) for 1 hat 37°C.
- Remove the coverslip and rinse 3 times at 2×SSC at room temperature (RT). Dehydrate in an ethanol series (70%, 85%, 95%) at RT and dry slide with compressed air.
- Incubate with 20 μl 0.4 ug/ml proteinase K in 20 mM Tris, pH 8.2, under a cover slip for 8-15 min at 37°C.
- Remove the cover slip and rinse in 2×SSC at RT.
- Fix in 4% paraformaldehyde in PBS with 50 mM MgCl, for 10 min at RT. Rinse slide 3 times in 2×SSC at RT.
- Dehydrate in ethanol series (70%, 85%, 95%) and dry with compressed air.
- Soak slide in 70% formamide in 2×SSC for 5 min at 70°C.
- Soak slide in 70% ethanol for 1 min at 4°C, dehydrate in ethanol series (70%, 85%, 95%), and dry with compressed air.
- Warm slide to 37°C and incubate for 16-24 h with 15 μl denatured (at 75°C for 5 min) and pre-annealed (at 37°C for 30 min) probe/hybridization solution [10.5 μl MMI, 30 ng labeled probe (3 μl), 3 ug mouse Cot-1 DNA (1.5 μl)] under a cover slip with rubber cement-sealed edges.
- Remove coverslip and wash 3 times in 50% formamide in 2X SSC for 2 min at 45°C, 3 times in 2×SSC, once in 0.1×SSC, and once in 0.1% TWEEN 20 for 2 min at RT. Block in PMN for 10 min at 4°C.
-
Browse our recommendations
Explore our scope support Transgenic Mapping (FISH) Services from sample preparation through to the final report. Creative Bioarray can use fluorescence in situ hybridization (FISH) to confirm the integration of transgenes and determine their chromosome band location. We guarantee the speed, quality, and cost of our service.
Detection of hybridization signal
- Incubate with 20 μl of 20 μg/ml sheep anti-digoxigenin in PMN in the dark for 45 min at RT or for 15-30 min at 37°C.
- Wash slides 3 times in PN buffer for 2 min at RT in the dark, drain, and dry back with a lint-free tissue.
- Cover with 15 μl propidium iodide in antifade solution under a cover slip and protect from light.
- Locate and photograph the same metaphase spreads that were photographed after G-banding.
NOTES
- Treatment of slides with the extremely low concentration of trypsin used for G-banding is not harmful to the structure of mouse chromosomes if the slides are appropriately fixed before denaturation for hybridization. Furthermore, treatment with trypsin does not affect the background.
- Sensitivity is increased with the use of high numerical aperture (>1.3) objectives.
RELATED PRODUCTS & SERVICES
Reference
- Shi YP, et al. (1994). "The mapping of transgenes by fluorescence in situ hybridization on G-banded mouse chromosomes." Mamm Genome. 5 (6), 337-341.