Neural Progenitor Cell Village Reveals Natural Variation
Cell Stem Cell. 2023 Feb 10; S1934-5909(23)00035-8.
Authors: Wells MF, Nemesh J, Ghosh S, Mitchell JM, Salick MR, Mello CJ, Meyer D, Pietilainen O, Piccioni F, Guss EJ, Raghunathan K, Tegtmeyer M, Hawes D, Neumann A, Worringer KA, Ho D, Kommineni S, Chan K, Peterson BK, Raymond JJ, Gold JT, Siekmann MT, Zuccaro E, Nehme R, Kaykas A, Eggan K, McCarroll SA.
INTRODUCTION
Humans harbor immense diversity in biological traits and disease risk, affecting almost all organs and physiological functions. Reservoirs of natural variation allow populations to adapt to existential crises and selective pressures, such as viral outbreaks. Genetic variants can influence neural phenotypes through the regulation of gene expression, which has unknown effects on signaling pathways, cell migration, and cell-cell interactions. Recognizing the underlying molecular and cellular mechanisms will require scalable approaches.
METHODS
- To ascertain how natural genetic variation shapes cellular phenotypes, we implemented a maximum likelihood approach that calculates for each donor the likelihood that the observed single-cell level combination of transcribed alleles arose from that donor's genome sequence. We provide the resulting software "Dropulation" in an open-source format.
- Constructing villages of NPCs requires quick, dependable induction, so we designed an approach to create and maintain self-renewing human NPCs. SW7388-1 iPSCs were transduced with two separate lentiviruses encoding TetO::Ngn2:T2A:PURO and Ubq::rtTA to enable doxycycline (DOX)-inducible expression of mouse NGN2 and the linked puromycin resistance gene. We initiated NGN2 induction by adding DOX and small molecule inhibitors of the SMAD and WNT signaling pathways, which dorsalizes early neural cells.
- The hPSC-to-NPC transition is accompanied by changes in molecular landscapes. We used qPCR, bulk RNA-seq, and ChIP-seq to evaluate SNaP transcriptional signatures. To assess developmental potency, we cultured SNaPs under conditions that promote differentiation into neuronal and glial lineages.
- To map expression QTLs (eQTLs) in human NPCs, we tested all genes for associations between expression levels and nearby SNPs among the donors in the SNaP-44 village using a linear regression model. Using SNaPs, CRISPR screens, and cell villages, we sought to identify genetic effects on a specific cellular phenotype-the vulnerability of human NPCs to the neurotropic ZIKV.
- We performed genome-wide CRISPR-Cas9 positive selection survival screens to determine which genes are important for ZIKV infection in SNaPs and to limit the number of potential genes that could explain variation in ZIKV susceptibility. Transduced SNaPs were treated with mock-infection media, ZIKV-Ug or ZIKV-PR.
RESULTS
- We constructed a 104-donor cell village that included whole-genome sequenced human induced pluripotent stem cells (iPSCs) derived from male and female skin and blood cells. The iPSCs from 104 donors exhibited highly similar RNA expression patterns. The primary source of variation in donor transcriptional profiles was their progress through the cell cycle. Consistent with earlier observations from tissue-level analysis, sex effects were small.
- At 48 h, the cells showed a bipolar morphology characteristic of NPCs. We passage into growth-factor-containing media, then we observed self-organized neural rosette structures and still-higher percentages of NPC marker positivity. SNaPs also displayed high expression of genes expressed in dorsal progenitors and low levels of genes expressed in posterior and ventral brain regions. SnaPs, therefore, resemble dorsal telencephalic NPCs. SNaPs were capable of self-renewal as depicted by clonal proliferation and differentiation assays.
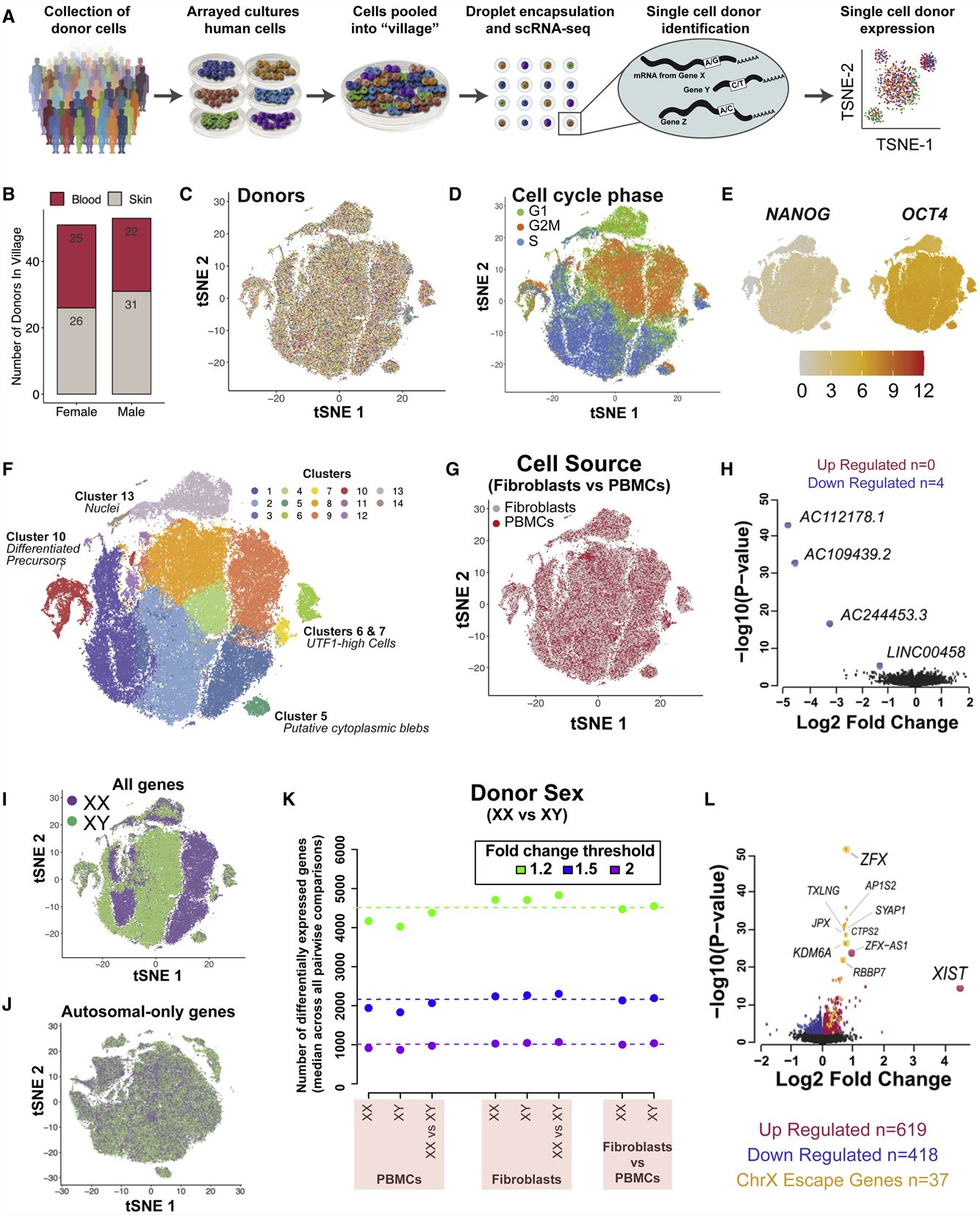
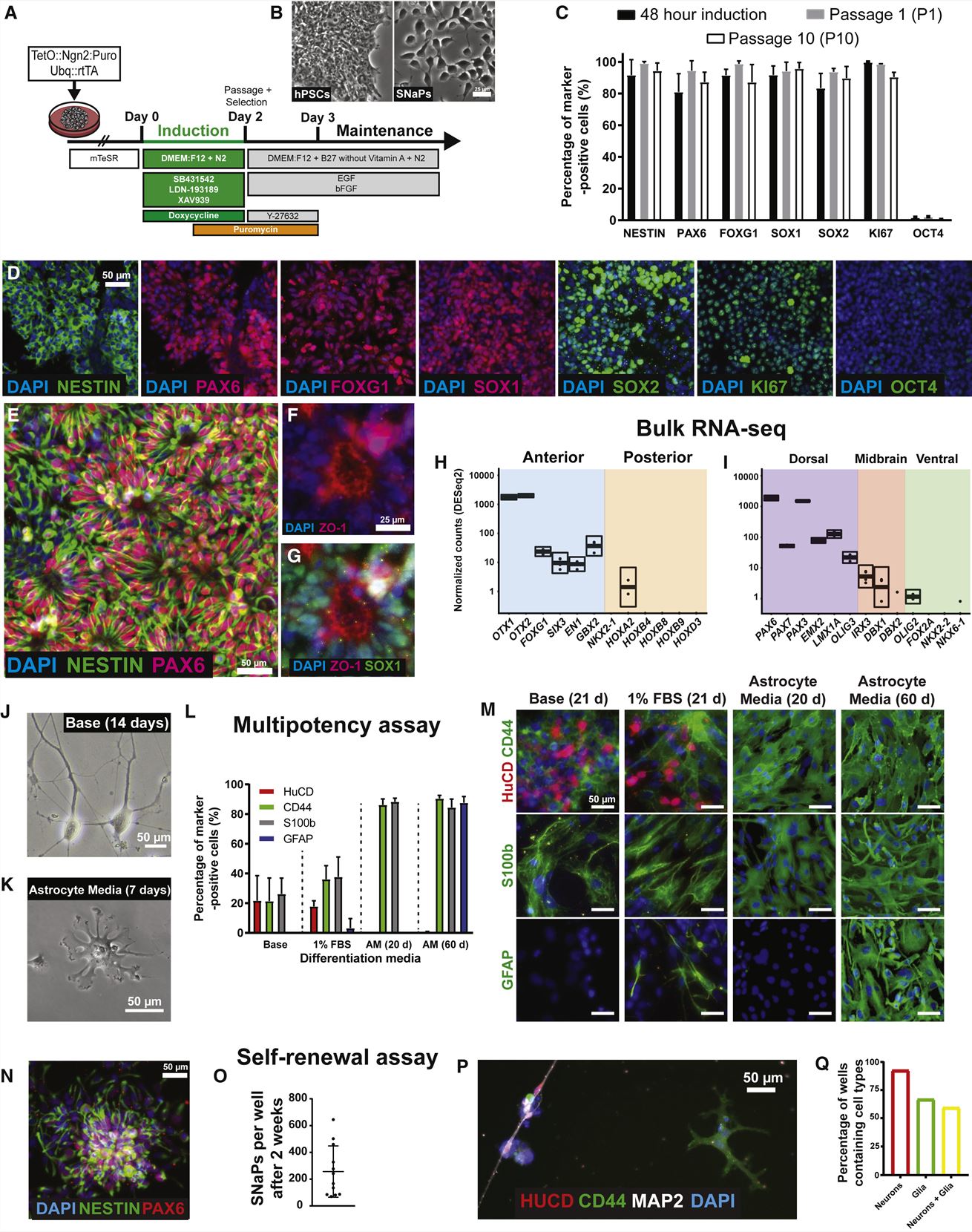
Fig. 1 Left: scRNA-seq characterization of human iPSC village; Right: Rapid induction of stem cell-derived human NPCs.
- Cell village eQTL analysis, therefore, uncovered genetic influencers of transcript levels in human NPCs that also affect neurodevelopmental phenotypes in vivo. We verified that SNaPs model ZIKV neuropathogenesis by immunostaining for ZIKV envelope protein (4G2). Several hits were confirmed through arrayed infectivity and viability assays performed in CRISPR-edited SNaPs that were generated from a constitutive-Cas9 H1 hESC line.
- SNaPs from donors homozygous for the reference allele (C) of rs34481144 expressed IFITM3 at levels 4.8-fold higher than donors homozygous for the alternate allele,and again found the same significant relationship between rs34481144 genotype and ZIKV infectivity. These results demonstrate that a genetically variable, cell-intrinsic property of human NPCs is the major source of inter-individual variation in their susceptibility to a viral pathogen.
- A significant increase in the size of CACHD1-edited organoids as early as day 9, when NPCs are the predominant cell type. CACHD1 editing drastically reduced the number of TBR1+ subplate neurons and TBR1+: SOX2+ cell ratio compared with NT gRNA controls, indicating significant defects in neurogenic potential. These findings highlight the importance of CACHD1 in neurogenesis while establishing SNaPs as an in vitro human model for future mechanistic studies of relevant disease mechanisms.
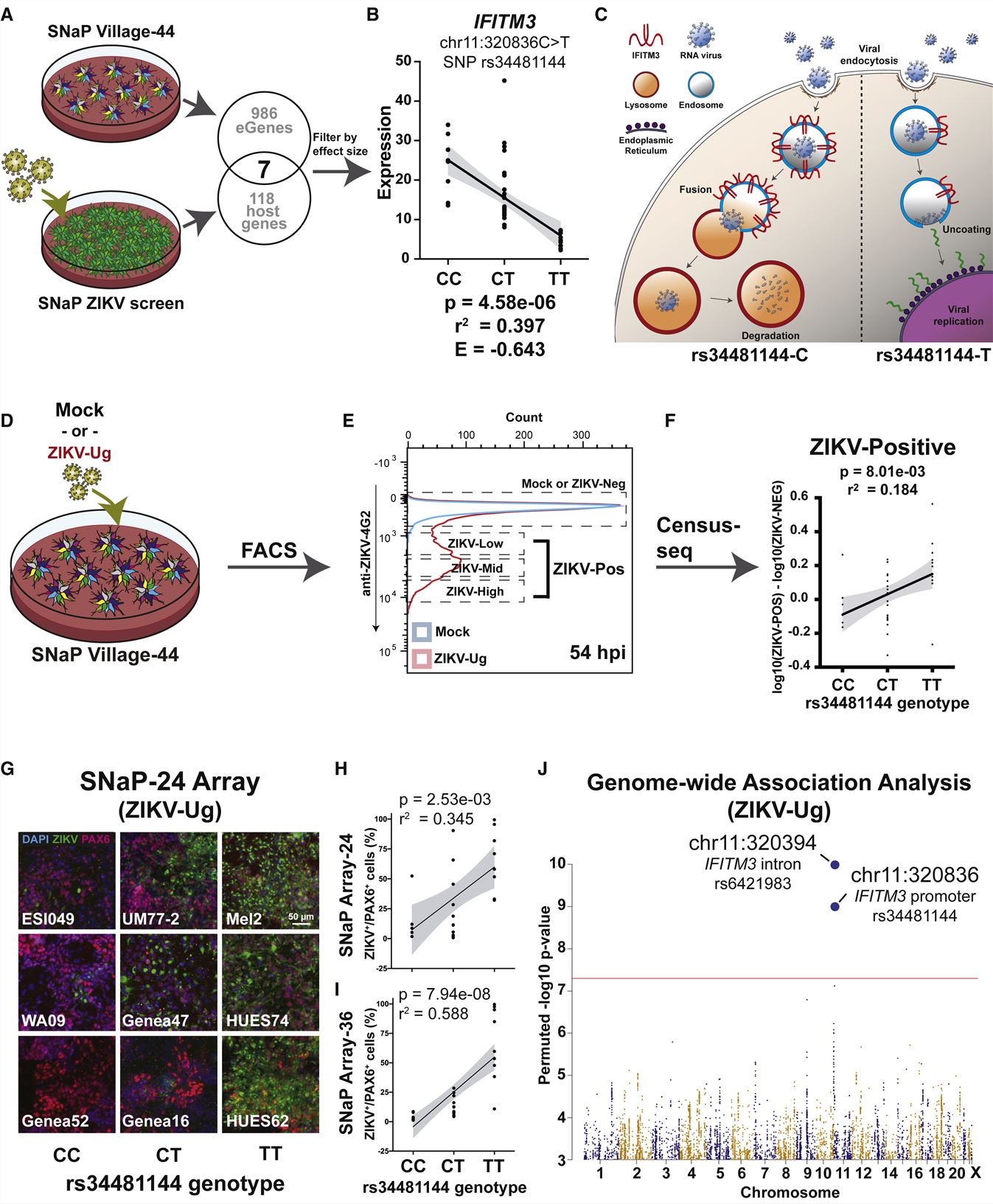
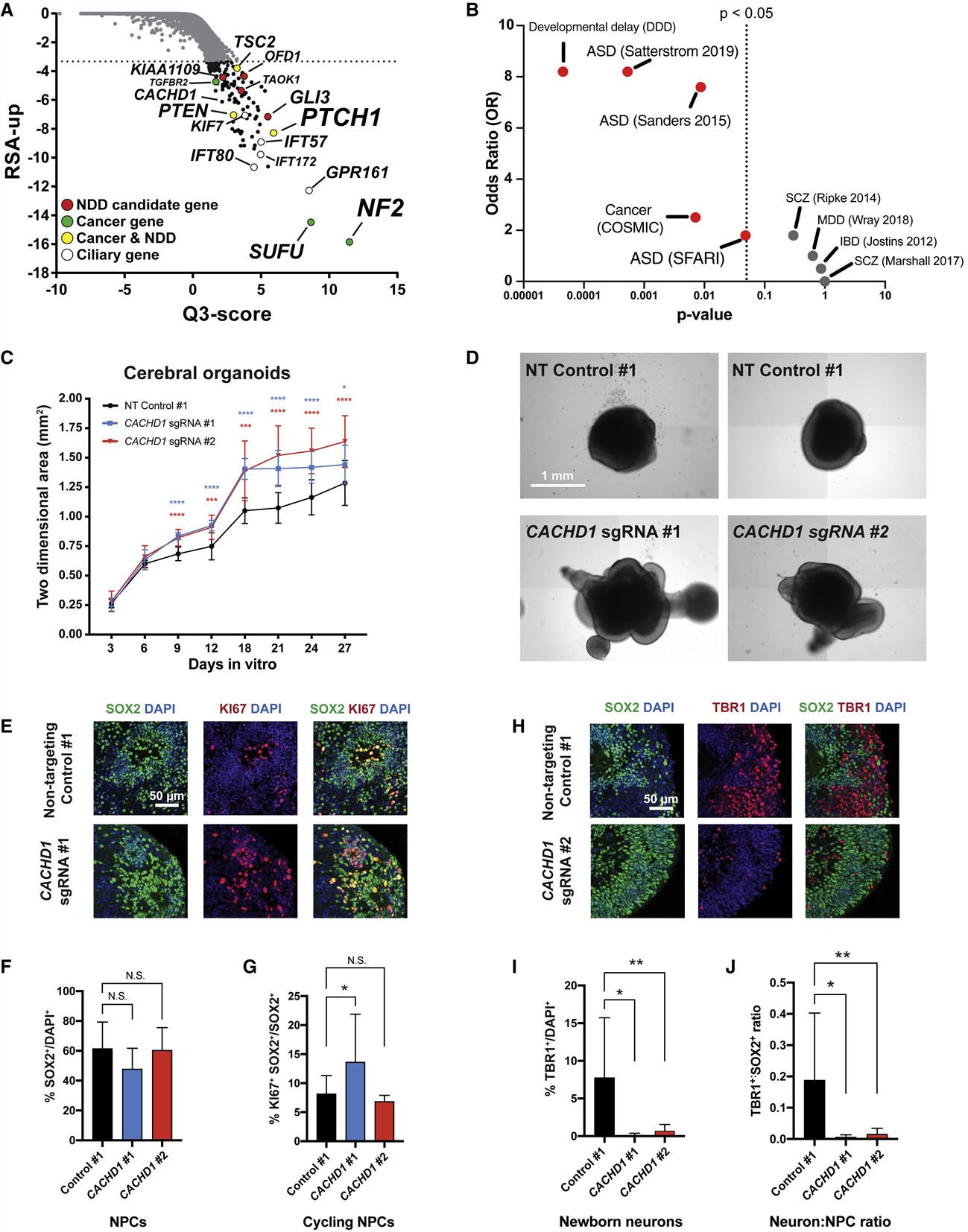
Fig. 2 Left: SNaP sensitivity to ZIKV associates to a common SNP in IFITM3. Right: CACHD1 regulates organoid neurogenesis.
SUMMARY
- Here, we describe a "cell village" experimental platform we used to analyze genetic, molecular, and phenotypic heterogeneity across neural progenitor cells.
- Through rapid induction of human stem cell-derived neural progenitor cells, measurements of natural genetic variation, and CRISPR-Cas9 genetic perturbations, we identified a common variant that regulates antiviral IFITM3 expression and explains most inter-individual variation in susceptibility to the Zika virus.
- We also detected expression QTLs corresponding to GWAS loci for brain traits and discovered novel disease-relevant regulators of progenitor proliferation and differentiation such as CACHD1.
RELATED PRODUCTS & SERVICES
Reference
- Wells MF, et al. (2023). "Natural variation in gene expression and viral susceptibility revealed by neural progenitor cell villages." Cell Stem Cell. S1934-5909(23)00035-8.