Multiscale 3D Genome Reorganization of Skeletal Muscle Stem Cells
Science Advances. 2023 Feb 17; 9 (7): eabo1360.
Authors: Zhao Y, Ding Y, He L, Zhou Q, Chen X, Li Y, Alfonsi MV, Wu Z, Sun H, Wang H.
INTRODUCTION
Skeletal muscle tissue has a remarkable regenerative capacity, which largely depends on the activation of resident muscle stem cells, named satellite cells (SCs). Little is known about three-dimensional (3D) genome organization in skeletal muscle stem cells. Recent advents in mapping genome-wide 3D chromosomal interactions have led to unprecedented understanding of hierarchical organization of the genome within the 3D nucleus and how gene expression is orchestrated at the 3D level.
METHODS
- To delineate the remodeling in genome architecture during SC lineage progression, SCs at various stages were prepared. We then probed 3D genome organization in the above cells by in situ Hi-C with four to seven biological replicates per sample.
- Next, we examined TAD organization during SC lineage development. To further investigate whether TAD reprogramming is correlated with the local gene expression, we found that principal components analysis (PCA) of IS kinetics showed a progressing trajectory. Leveraging the H3K27ac chromatin immunoprecipitation sequencing (ChIP-seq) profiles in QSC and FISC, we defined SEs by ROSE algorithm, and the remaining enhancers were automatically classified into typical enhancers (TEs).
- To further investigate the possible regulatory role of PAX7 in E/P loop formation, we analyzed the publicly available PAX7 ChIP-seq profile from induced muscle progenitors. To further elucidate the promoting role of PAX7 binding on loop formation, we deleted Pax7 in QSCs by exploiting our recently developed muscle-specific CRISPR-Cas9/adeno-associated virus (AAV)-single guide RNA (sgRNA) in vivo genome editing system. We isolated QSCs from the mice 4 weeks after virus injection and performed Hi-C analysis.
- We thus took a close examination at the 3D organization associated with the Pax7 locus. To further dissect key elements governing the above-observed 3D reorganization, we used the looping interactions as well as assay for transposase-accessible chromatin using sequencing (ATAC-seq; from FISC) and H3K27ac ChIP-seq (from both QSC and FISC) signals. Last, to further probe into the mechanism of RE regulation of Pax7 expression, we searched for putative TF motifs in each RE region.
- To delineate the changes in 3D genome organization with SC aging, we performed in situ Hi-C at the above stages with three to four replicates for each to obtain a total of >300 million uniquely aligned contacts.
RESULTS
- We obtained a resolution of 5 kb for QSC and FISC, and 10 kb for D1, D2, and D3 after in-depth sequencing, which rendered us to explore the 3D genome architecture at multiscale, including compartments, TADs, and chromatin loops. Genome compartmentalization is dynamically rewired during SC lineage progression and most pronounced when cells become activated; compartmentation remodeling proceeds expression changes in a substantial number of genes.
- Aggregate interaction matrices also revealed a genuine loss of insulation across the TAD borders early in FISC versus QSC. IS also increased drastically in FISC versus QSC but decreased when SCs were activated, proliferated, and differentiated. The above findings demonstrate that border reorganization occurs during the entire lineage development. Unexpectedly, IS increase did not appear to proceed the transcriptional changes; concomitant changes were observed on many loci. In addition, the presence of SE clusters was associated with more evident expression changes; transcription levels in SE-containing TAD clusters in both QSC and FISC displayed stage-specific expression patterns.
- We found that, although enhancer and promoter landscapes were largely unaltered in FISC versus QSC, the loop formation was highly dynamic, with 95.2 and 64.3% specifically identified in QSC and FISC, respectively. In addition, stage-specific E-P and E-E loops also displayed more prominent difference in APA scores and correlation with gene expression. PAX7 binding was enriched at the loop anchors of QSC-specific loops and that the number of PAX7-bound E/P loops (PAX7+ E/P loops) sharply decreased in FISC versus QSC. The PAX7 KD caused a loss in the total number of chromatin loops, and those E/P loops with PAX7 binding also showed notable decrease of interaction strength.
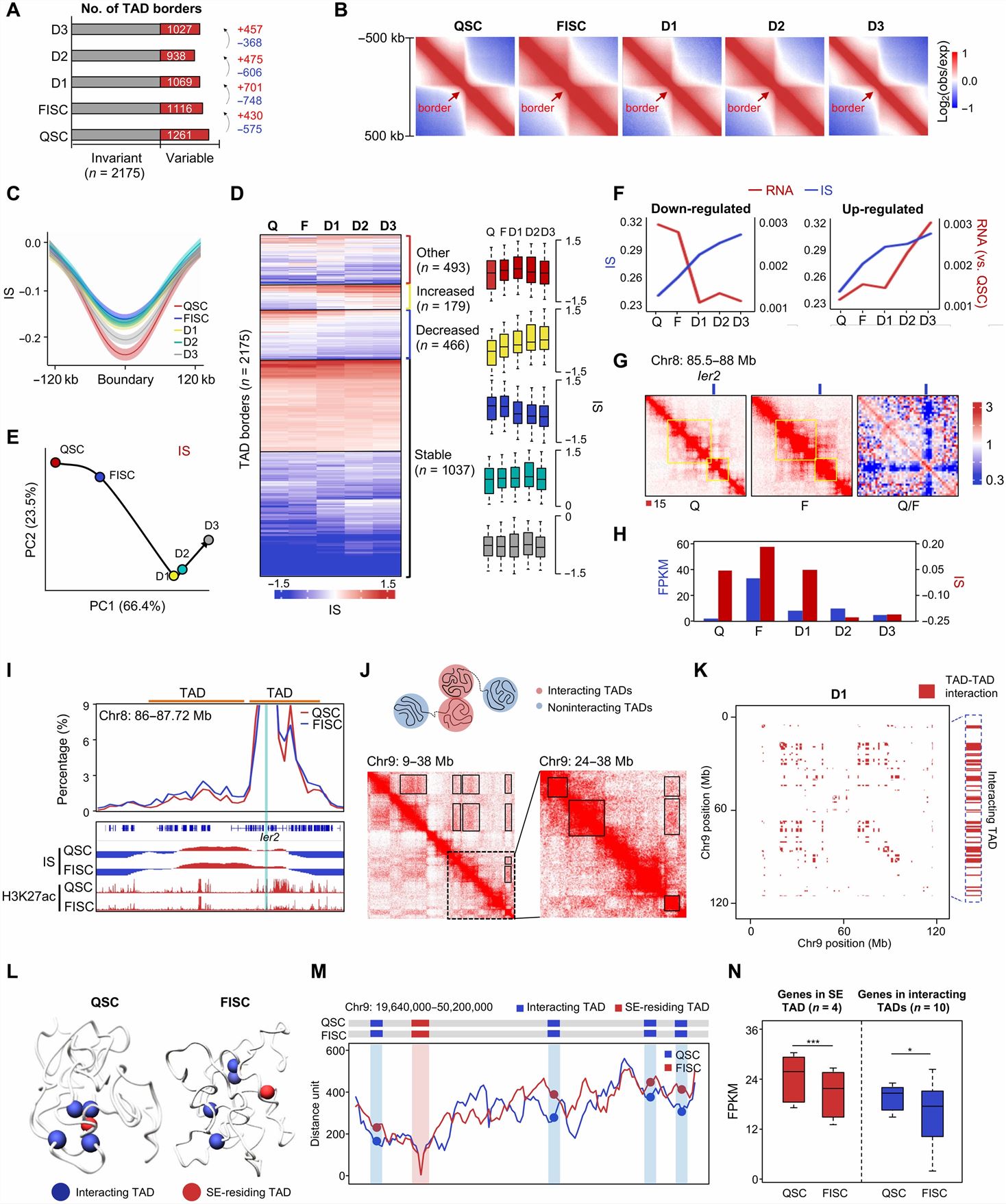
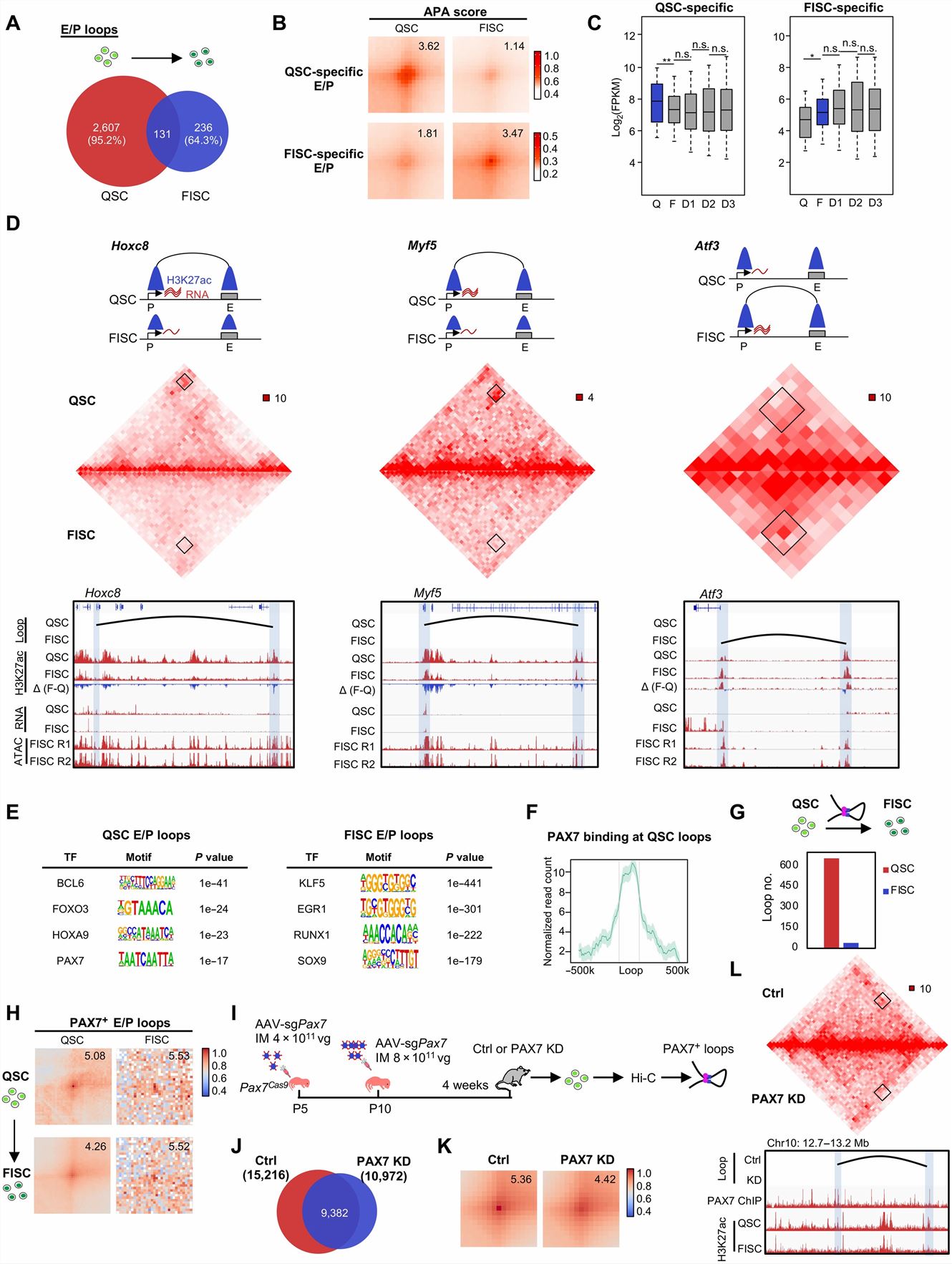
Fig. 1 Left: Dynamic TAD organization during SC lineage development; Right: PAX7 regulates E/P loop reorganization during SC early activation.
- Pax7 locus undergoes drastic reorganization at the 3D level when SCs become activated, and this may account for its expression attenuation. We finally identified four putative REs (R1 to R4), there are evident loops among R1/R4, R2/R4, and R3/R4 in QSC, but only one R1/R4 loop retained in FISC; R2, R3, or R4 deletions caused a much less pronounced Pax7 decrease in FISC. At the loop level, these RE-anchored intra-TAD loops were largely attenuated or even lost upon RE removal. PAX7 KD caused a significant increase of the inter-Pax7 TAD interaction strength.
- Young SCs exhibited distinct 3D genome organization compared with aged or geriatric SCs; long-range (>3 Mb) interaction frequencies increased significantly in geriatric SCs compared with young and aged SCs. We found an increase in compartmentalization (decreased intercompartment contacts) at aged versus young SCs, but then reduced at geriatric stage. TAD borders largely lost insulation strength when SCs entered into geriatric state. Furthermore, we found that local gene expression underwent a significant increase or decrease when TAD borders were gained or lost, respectively. In addition, the DS kinetics was highly correlated with changes in PC1 values or gene expression during the transition between two stages.
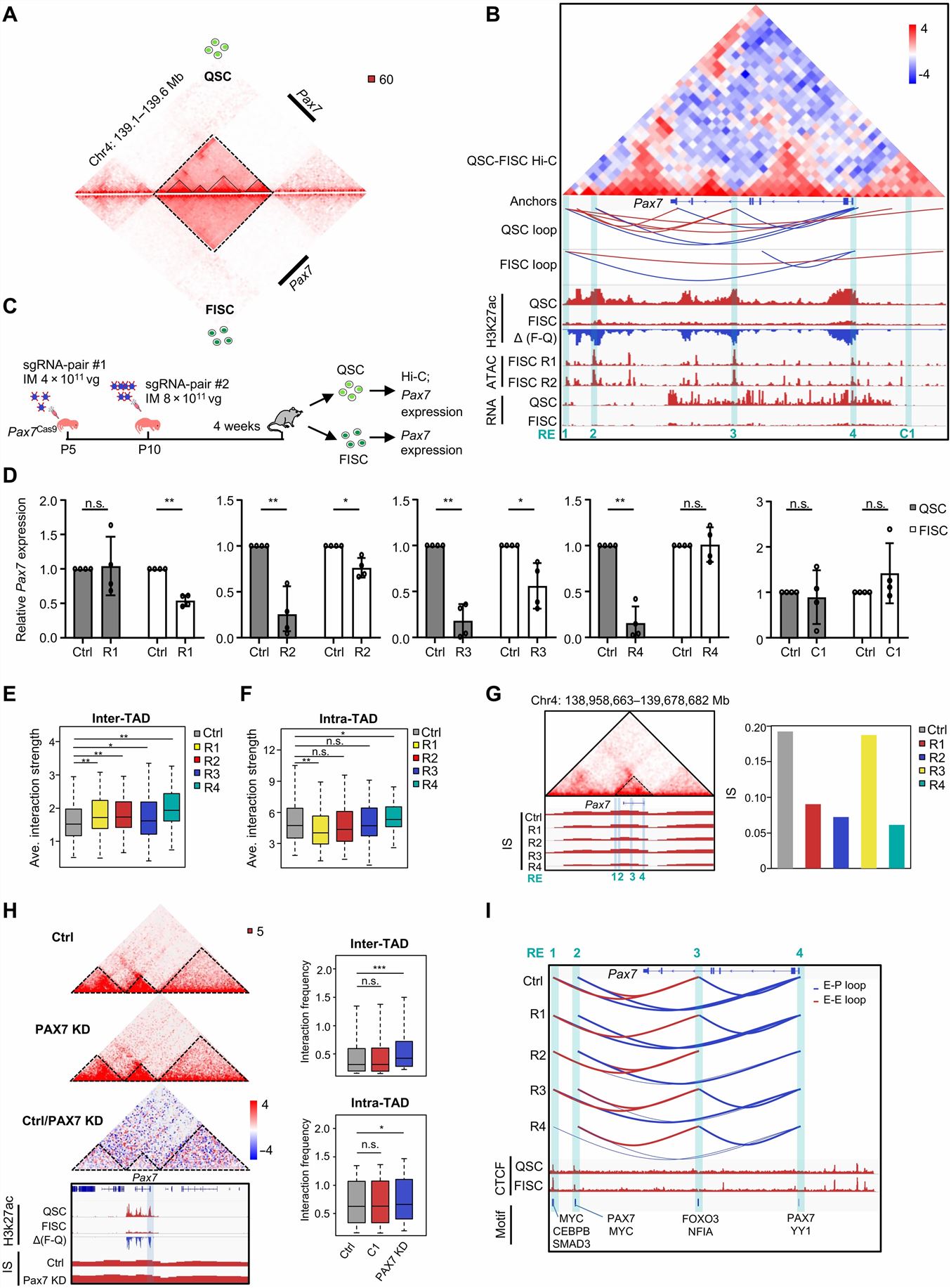
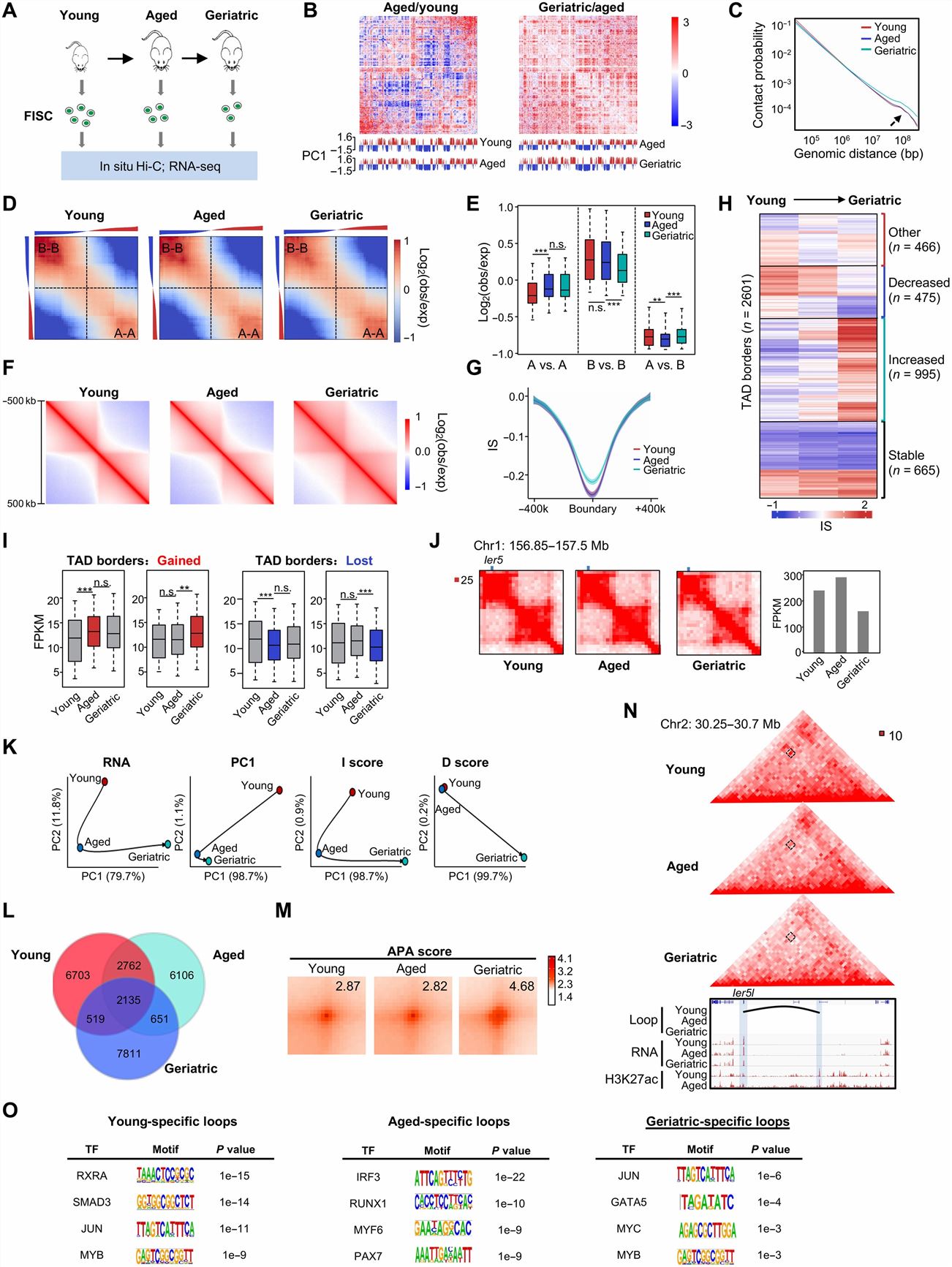
Fig. 2 Left: 3D regulatory interactions orchestrate Pax7 expression dynamics in SC; Right: Multiscale analysis of 3D genome during mouse SC aging.
SUMMARY
- Here, we comprehensively map the 3D genome topology reorganization during mouse SC lineage progression. Our results uncover that 3D chromatin extensively reorganizes at multiple architectural levels and underpins the transcriptome remodeling during SC lineage development and SC aging.
- Rewiring at the compartment level is most pronounced when SCs become activated. Marked loss in topologically associating domain (TAD) border insulation and chromatin looping also occurs during early activation process. Meanwhile, TADs can form TAD clusters and super-enhancer-containing TAD clusters orchestrate stage-specific gene expression.
- We uncover that transcription factor PAX7 is pivotal in enhancer-promoter (E-P) loop formation. We also identify cis-regulatory elements that are crucial for local chromatin organization at Pax7 locus and Pax7 expression. Lastly, we unveil that geriatric SC displays a prominent gain in long-range contacts and loss of TAD border insulation.
RELATED PRODUCTS & SERVICES
Reference
- Zhao Y, et al. (2023). "Multiscale 3D genome reorganization during skeletal muscle stem cell lineage progression and aging." Sci Adv. 9 (7), eabo1360.