TUNEL Apoptosis Assay (Fluorescent)
Introduction
The mechanisms of apoptosis can represent a critical aspect of toxicological analysis and drug discovery. DNA fragmentation is a hallmark of late stage apoptosis, which can be detected in fixed cells and tissue sections by the terminal deoxynucleotidyl transferase (TdT) mediated dUTP nick-end labeling (TUNEL) assay.
The TUNEL assay relies on the TdT enzyme to catalyze the addition of labeled dUTP (BrdUTP or EdUTP) to the 3'-hydroxyl (3'-OH) ends of cleaved DNA fragmentations. Fluorescent dye-conjugated dUTP can be used for direct detection of fragmented DNA by fluorescence microscopy or flow cytometry. It is highly selective for the detection of apoptotic cells but not necrotic cells or cells with DNA strand breaks resulting from irradiation or drug treatment.
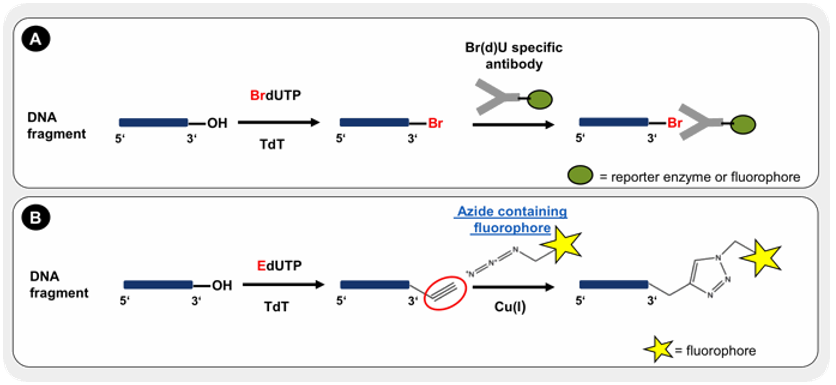 Figure 1. The principle of TUNEL assay. Incorporated BrdUTP is detected by specific antibody conjugates with a reporter enzyme or fluorescent dye (A). The incorporation of EdUTP is visualized by Cu-catalyzed alkyne-azide click chemistry with an azide containing fluorphore (B).
Figure 1. The principle of TUNEL assay. Incorporated BrdUTP is detected by specific antibody conjugates with a reporter enzyme or fluorescent dye (A). The incorporation of EdUTP is visualized by Cu-catalyzed alkyne-azide click chemistry with an azide containing fluorphore (B).
Materials
TdT reaction buffer
EdUTP nucleotide mixture
TdT recombinant
DNase I
DNase I buffer
Staining buffer
Fluor picolyl azide dye
Hoechst 33342
1X phosphate buffered saline (PBS)
4% paraformaldehyde in PBS (fixative)
0.25% Triton X-100 in PBS (permeabilization reagent)
3% bovine serum albumin (BSA) in PBS
96-well plate (for the specific, automated imaging instrument)
22 × 22 mm or 18 × 18 mm coverslips (for standard microscopy)
Protocol Summary
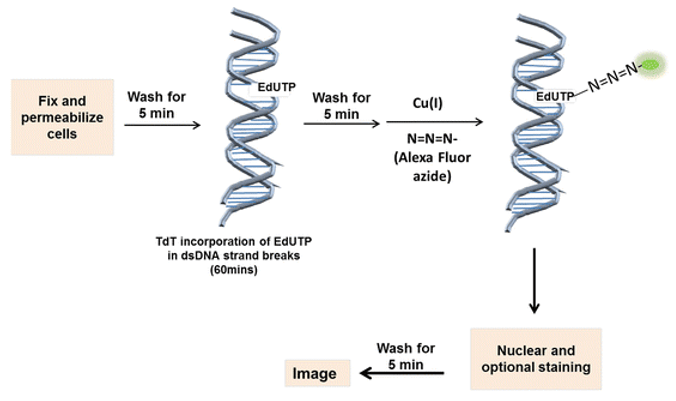
Cell Fixation and Permeabilization
This protocol is amenable to other fixation and permeabilization reagents such as 70% ethanol.
- Remove media and wash 96-well plate or coverslips once with PBS.
Note: If there is a chance that cells lost by this step, we recommend fixing directly without performing the wash steps. - Add a sufficient volume of fixative (4% paraformaldehyde) to completely cover the samples.
- Incubate for 15 minutes at room temperature.
- Add sufficient volume of the permeabilization reagent (0.25% Triton X-100 in PBS) to completely cover the samples.
- Incubate samples for 20 minutes at room temperature.
- Wash twice with deionized water.
Positive Control Preparation
The DNase I induces strand breaks in the DNA to provide a positive TUNEL reaction.
- Prepare DNase I solution according to Table 1 and mix well.
Note: Do not vortex the DNase I solution as DNase I denatures with vigorous mixing. - Add 100 μL of the DNase I solution to each sample and incubate for 30 minutes at room temperature.
- Wash once with deionized water, proceed to TUNEL reaction.
Table 1. DNase I solution.
Reaction components Number of Samples 1 2 3 Deionized water 89 µL 178 µL 267 µL DNase I buffer 10 µL 20 µL 30 µL DNase I 1 µL 2 µL 3 µL Total volume 100 µL 200 µL 300 µL
TUNEL Reaction
- Add 100 μL TdT reaction buffer to each sample and allow the solution to spread completely over the surface.
- Incubate for 10 minutes at room temperature.
- Prepare the TdT reaction cocktail according to Table 2.
- Add 100 μL of the TdT reaction cocktail and allow the solution to spread completely over the surface.
- Incubate for 60 minutes at 37°C.
Note: Use a humidified chamber to protect against evaporation and this reaction can be performed overnight at room temperature. - Wash samples twice with 3% BSA in PBS.
Table 2. TdT reaction cocktails.
Reaction components Number of Samples 1 2 4 5 10 TdT reaction buffer 94 µL 188 µL 376 µL 470 µL 940 µL TdT recombinant 4 µL 8 µL 16 µL 20 µL 40 µL EdUTP 2 µL 4 µL 8 µL 10 µL 20 µL Total volume 100 µL 200 µL 400 µL 500 µL 1 mL - Prepare the fluorescent staining solution according to Table 3 and mix well.
- Immediately add 100 μL of the fluorescent staining solution to each sample and allow the solution to spread completely over the surface.
Table 3. Fluorescent staining cocktails.
Reaction components Number of Samples 1 2 4 5 8 12 100 Staining buffer 97.5 µL 195 µL 390 µL 487.5 µL 780 µL 1.17 mL 9.75 mL Fluor picolyl azide dye 2.5 µL 2 µL 10 µL 12.5 µL 20 µL 30 µL 250 µL Total volume 100 µL 200 µL 400 µL 500 µL 800 µL 1.2 mL 10 mL - Incubate for 30 minutes at room temperature, protected from light.
- Wash coverslips or 96-well plate twice with 3% BSA in PBS for 5 minutes.
DNA Staining
- Dilute Hoechst 33342 in PBS to obtain a 1 X Hoechst 33342 solution.
- Add 100 μL 1 X Hoechst 33342 solution per coverslip or well
- Incubate for 15 minutes at room temperature, protected from light.
- Wash each coverslip or well twice with PBS.
Imaging and Analysis
Image plate or coverslip using filters appropriate for the fluorescent dye.
Table 4. Approximate fluorescence excitation/emission (Ex/Em) maxima.
| Fluorophore | Excitation (nm) | Emission (nm) |
| Fluor 488 | 495 | 519 |
| Fluor 594 | 590 | 615 |
| Fluor 647 | 650 | 670 |
| Hoechst 33342 bound to DNA | 350 | 460 |
Cell Services:
Cell Line Testing and Assays: