Bivariate Cell Cycle Assay (Cyclins/PI)
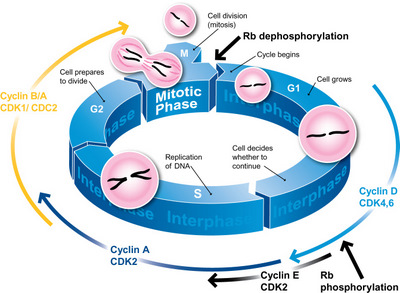
The cell cycle is the most basic and important process in eukaryotic cells. Being an ordered set of events, ending up with cell growth and division into two daughter cells, the cell cycle is tightly regulated by defined temporal and spatial expression, localization, and destruction of several cell cycle regulators. Cyclins are the major control switches of the cell cycle progression. In particular, the expression of cyclins D, E, A, and B1 provides a new cell cycle landmarks that can be used to subdivide the cell cycle into several distinct sub-compartments.
This approach below is based on the bivariate analysis of DNA content and cyclins, allowing differentiation between G0 and G1 cells, identification of mitotic cells, and measurement relate expression of other intracellular proteins to the cell cycle position.
The cell cycle assays are applicable to many areas of life science research and drug development, including cancer biology, apoptosis analysis, drug screening, and measuring health status of cell cultures in bioreactors.
Materials
Ethanol
Triton X-100
Tris-EDTA buffer
Anti-cyclins mAbs
Propidium iodide (PI)
Paraformaldehyde (PFA)
Goat-anti-mouse-FITC
Phosphate buffered saline (PBS)
Fixation
For cyclins E, A, and B1, cells (2 x 106/mL) are fixed by 70% ethanol:
- Put all the reagents on ice.
- Count and centrifuge the cells for 5 min at 300 g.
- Resuspend the pellet in 1mL Tris-EDTA buffer.
- Add 3 mL of 95% ethanol, vortex gently on ice.
- Store the fixed samples at 4°C.
In the case of cyclin D, cells are fixed with 1% methanol-free formaldehyde:
- Put all the reagents on ice.
- Count and centrifuge the cells at 300 g for 5 min.
- Suspend the pellet in 1 mL of 1% formaldehyde in PBS for 15 min on ice.
- Wash with ice-cold PBS and centrifuge at 300 g for 5 min.
- Add 1 mL of 70% ethanol.
- Store the fixed samples at 4°C.
Staining Procedure
- Wash with ice-cold PBS.
- Resuspend the pellet in 1 mL of 0.25% Triton X-100 and vortex samples gently on ice (for 5 min in the case of cyclins D, A, and B1; for 10-20 min in the case of cyclin E).
- Wash with ice-cold PBS and centrifuge at 300 g for 5 min.
- Incubate cells with 150 μL of anti-cyclin mAb (diluited at the concentration of 2.5 μg/mL in PBS) overnight at 4°C.
- Wash with ice-cold PBS and centrifuge at 300 g for 5 min.
- Incubate for 1 hour with 150 μL secondary mAb goat-anti-mouse-FITC (diluited 1:50 in PBS).
- Wash with ice-cold PBS and centrifuge at 300 g for 5 min.
- Add 1 mL of 2.5 μg/mL PI (containing RNase) and incubate for 4 hours at room temperature or add 0.8 μg/mL PI (containing RNase) and incubate overnight at 4°C.
- Analyse with flow cytometer equipped with a 488 nm argon laser.
Note: 1) The key steps in this method are cell fixation, permeabilization, and the concentrations of anti-cyclin mAbs. For most cyclins, the optimal fixative is 70% ethanol. This treatment preserves cyclins, reducing the background and non-specific fluorescence, and resulting in an improved signal-to-noise ratio of the cyclin-specific fluorescence, while cyclin D requires fixation in formaldehyde. As far as the concentration of anti-cyclin mAb, 2.5 μg/mL is optimal for most cells. 2) In this procedure, a negative control sample containing only the secondary mAb-FITC is required.
References
- Gong J, et al.; Simultaneous analysis of cell cycle kinetics at two different DNA ploidy levels based on DNA content and cyclin b measurements. Cancer Res, 1993, 53: 5096.
- Gong J, et al.; Discrimination of G2 and mitotic cells by flow cytometry based on different expression of cyclins A and B1. Exp Cell Res, 1995, 220: 226.
- Widrow R. J. et al.; Separation of cells at different times within G2 and mitosis by cyclin B1 flow cytometry. Cytometry, 1997, 27: 250.
- Darzynkiewicz Z, et al.; Cytometry of cyclin proteins. Cytometry, 1996, 25: 1.
Cell Services:
Cell Line Testing and Assays: