Application of aCGH for Identification of Chromosomal Aberrations
Journal of Assisted Reproduction and Genetics. 2022 Feb; 39 (2): 357-367.
Authors: Kowalczyk K, Smyk M, Bartnik-Głaska M, Plaskota I, Wiśniowiecka-Kowalnik B, Bernaciak J, Chojnacka M, Paczkowska M, Niemiec M, Dutkiewicz D, Kozar A, Magdziak R, Krawczyk W, Pietras G, Michalak E, Klepacka T, Obersztyn E, Bal J, Nowakowska BA.
INTRODUCTION
- Before the era of molecular cytogenetics, a routine karyotype analysis by GTG technique was the only method used to detect trisomy and monosomy of whole chromosomes or deletions and duplications greater than 5-10 Mb in size. Over the past several years, new techniques for fast diagnosis of the most common chromosomal aneuploidy in the fetus were introduced.
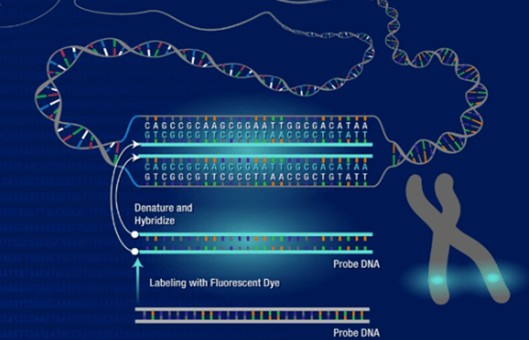
- These techniques include the following, Rapid-FISH (rapid fluorescent in situ hybridization), QF-PCR (quantitative fluorescent polymerase chain reaction), BoBs (BACs-on beads), MLPA (multiplex ligation-dependent probe amplification) technique, and CGH microarray (comparative genomic hybridization). For the next few years, it was proven that the aCGH (array-CGH) is the most effective and quickest method for detecting chromosomal aberrations in the material of miscarriage.
METHODS
- Sample types and DNA isolation. Forty-seven trophoblast and 15 fetal skin fibroblast samples were used for our study. Genomic DNA was extracted using the DNA isolation kit. For trophoblast and skin fibroblasts, incubation at 56°C with 20 µl proteinase K, water, and tissue lysis buffer was performed for at least 1 h for efficient digestion and lysis of the complete sample.
- Genomic array platform (array CGH analysis and interpretation). Array CGH was performed using 4 × 180 K microarrays. The array used in this study contains 60-mer oligonucleotide probes covering the whole genome with an average spatial resolution of 24 kb. All genomic coordinates are based on the reference genome (NCBI37/hg19). Data analysis and results were then performed with CytoSure Interpret Software. Quality control metrics are monitored with CytoSure Interpret Software.
- CNV classification. The study used five categories of classification of results, pathogenic, likely pathogenic, variants of unknown significance (VOUS), likely benign, and benign.
- Rapid-FISH. This method is performed on interphase nuclei immediately after collecting a small amount of trophoblast, which allows obtaining the result of a genetic test within 2-5 days. We used a commercially available set of probes containing centromeric probes for X, Y, 16, and 18 chromosomes and probes specific for critical regions of chromosomes 13, 21, and 22. For each preparation, at least 30 interphase nuclei were analyzed.
- Browse our recommendations
As an established leader in the field of cell science, Creative Bioarray offers a range of high-quality products and services for our customers' research, including but not limited to the products in the table below.
| Product/Service Types | Description | Recommended Products |
| Nucleic Acid Extraction | Nucleic acid extraction is the basic method and plays a vital role in molecular biology as the primary step for many downstream applications. | DNA Extraction Kits, RNA Extraction Kits, RNA/DNA Extraction Kits… |
| FISH | FISH is a cytogenetic technique using fluorescent probes to bind the chromosome with a high degree of complementarity. | Fluorescent In Situ Hybridization (FISH) Service, FISH Probe Design, Synthesis, and Testing Service, FFPE FISH Pretreatment Kit… |
| Molecular Karyotyping (aCGH) Service | Array Comparative Genomic Hybridization (aCGH) is a high-resolution karyotype analysis solution. | Molecular Karyotyping (aCGH) Service |
RESULTS
- Normal aCGH results were obtained in 27 samples (43.5%). In 22 female fetuses with normal CGH results, Rapid-FISH was carried out to exclude chromosomal polyploidy. In 3 cases (3/22, 13.6%) with the normal aCGH test result, the Rapid-FISH test showed chromosomal triploidy. One patient had a normal aCGH result, while Rapid-FISH found mosaic trisomy of chromosome 13 in 9% of cells (1/22, 4.5%).
- Abnormal aCGH and Rapid-FISH results were found in 35 cases (56.5% of cases). Chromosomal aneuploidy was detected in 23 cases. Pathogenic structural aberrations were found in 3 cases and are likely pathogenic in 3 cases. Aberrations responsible for miscarriage include the following: deletion 7p22.3p12.3 and duplication 9p24.3p13.2 (derived from parental balanced translocation), deletion 3q13.31q22.2 and deletion 3q22.3q23, and deletion 17p13.1 and deletion 1q21.1q21.2.
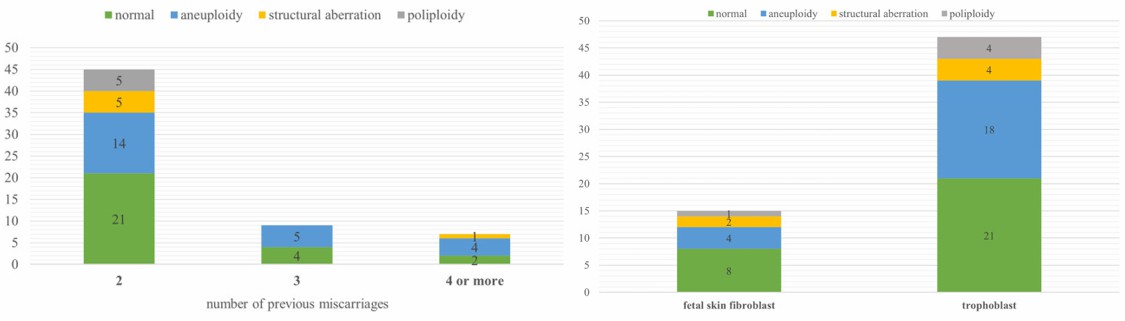 Fig. 1 Left: Number of cases with normal results, aneuploidy, polyploidy, and structural aberration in three subgroups of different numbers of previous miscarriages; Right: Number of cases with normal results, aneuploidy, polyploidy, and structural aberration in two subgroups of different material for DNA extraction.
Fig. 1 Left: Number of cases with normal results, aneuploidy, polyploidy, and structural aberration in three subgroups of different numbers of previous miscarriages; Right: Number of cases with normal results, aneuploidy, polyploidy, and structural aberration in two subgroups of different material for DNA extraction.
SUMMARY
- The research has proven that the method of comparative genomic hybridization to microarray is effective in identifying the most common genetic aberrations, submicroscopic genomic rearrangements, as well as genes whose mutations contribute to miscarriages.
- Of the 62-examined trophoblast/fetal skin fibroblasts, we found chromosomal aberrations in 56.5% of cases, and 82.9% of abnormal results were chromosomal numerical aberrations (trisomies, X-chromosome monosomy, and polyploidy), but as many as 17.1% were small aberrations that cannot be identified by the classical method GTG.
RELATED PRODUCTS & SERVICES
Reference
- Kowalczyk K, et al. (2022). "Application of array comparative genomic hybridization (aCGH) for identification of chromosomal aberrations in the recurrent pregnancy loss." J Assist Reprod Genet. 39 (2), 357-367.