Biofilm Imaging with FISH
Frontiers in Cellular Infection Microbiology. 2023 May 22; 13: 1195803.
Authors: Barbosa A, Miranda S, Azevedo NF, Cerqueira L, Azevedo AS
INTRODUCTION
Biofilms are complex structures with an intricate relationship between the resident microorganisms, the extracellular matrix, and the surrounding environment. Interest in biofilms is growing exponentially given their ubiquity in so diverse fields such as healthcare, environment, and industry. Molecular techniques (e.g., next-generation sequencing, RNA-seq) have been used to study biofilm properties. However, these techniques disrupt the spatial structure of biofilms; therefore, they do not allow us to observe the location/position of biofilm components (e.g., cells, genes, metabolites), which is particularly relevant to exploring and studying the interactions and functions of microorganisms.
Fluorescence in situ hybridization (FISH) has been arguably the most widely used method for an in situ analysis of spatial distribution of biofilms. In this review, an overview of different FISH variants already applied in biofilm studies (e.g., CLASI-FISH, BONCAT-FISH, HiPR-FISH, seq-FISH) will be explored.
FISH-Based Techniques Applied to Biofilms
In fact, in biofilm studies, the cells are not displayed in one layer but embedded in a 3D extracellular matrix. The bottom cells are usually less active (due to their inability to receive all the nutrients and oxygen that they need to survive), which implies fewer copies of rRNA and a lower signal intensity that might not be easily observable by epifluorescence microscopy. Hence, catalyzed reporter deposition (CARD)-FISH and nucleic acid mimics-FISH (NAM-FISH) have emerged to improve the performance of FISH for the in situ visualization of biofilms. On the other hand, another concern in the study of biofilm dynamics is that the spatial organization and composition of biofilm evolves. As such, fluorescence in vivo hybridization (FIVH) has tackled this challenge by providing in vivo information on the changes occurring in the biofilm.
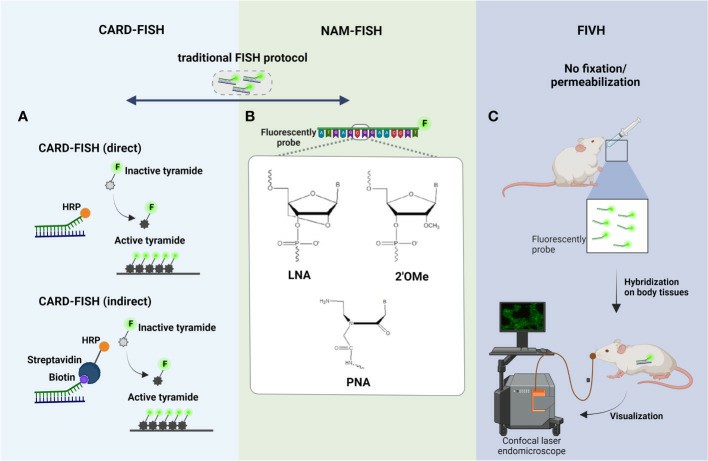 Fig. 1 Principal steps of (A) CARD-FISH, (B) NAM-FISH, and (C) FIVH.
Fig. 1 Principal steps of (A) CARD-FISH, (B) NAM-FISH, and (C) FIVH.
- CARD-FISH. There were developed two different variants of CARD-FISH: a direct method using probes directly linked with horseradish peroxidase (HRP) and an indirect method using biotinylated probes and HRP-labeled streptavidin. CARD-FISH has been shown to increase the sensitivity 26- to 41- times more than standard FISH.
- NAM-FISH. The chemical modifications of NAMs might be at nucleobase, sugar ring, or phosphodiester backbone level. Nowadays, there are available different NAM probes, but the most used are the Peptide Nucleic Acid (PNA), Locked Nucleic Acid (LNA), and 2'-O-methyl-RNA (2'OMe).
- FIVH. FIVH emerged to efficiently detect microorganisms inside the human body or the body of other higher-order animals. The LNA/2'OMe probe used in the FIVH protocol has not only worked efficiently at 37°C without toxic chemical compounds but also in the presence of gastric juice and low pH, as observed in stomach mucosa.
Multiplex FISH-Based Techniques Applied on Multispecies Biofilms Studies
Although, the study of single-species biofilms is more comprehensive, bacterial social interactions in multispecies biofilms have been gathering scientific interest and therefore multispecies studies in biofilms are increasing. As so, multiplexed FISH methodologies, including double-labeling-of-oligonucleotide (DOPE)-FISH and combinatorial labeling and spectral imaging (CLASI)-FISH, emerged to increase the number of different species/targets detected in a unique biofilm sample.
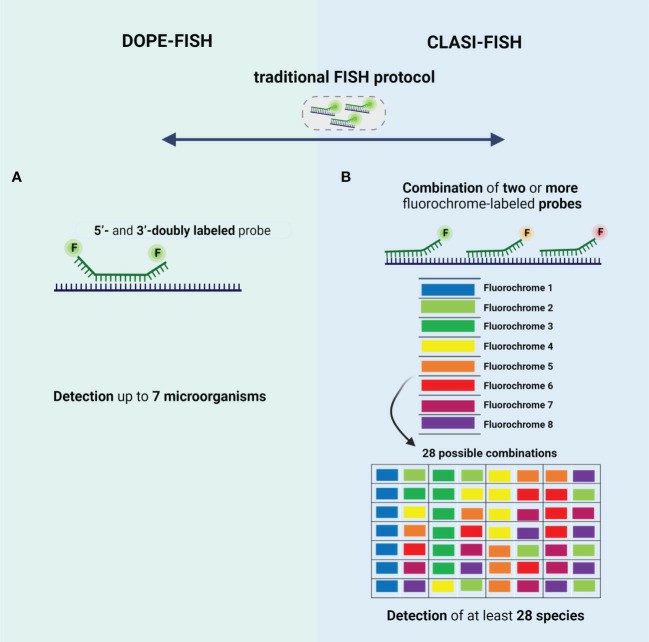 Fig. 2 Schematic representation of the type of probes used in (A) DOPE-FISH and (B) CLASI-FISH.
Fig. 2 Schematic representation of the type of probes used in (A) DOPE-FISH and (B) CLASI-FISH.
- DOPE-FISH. DOPE-FISH is a straightforward FISH variant that allows to detection of up to six microorganisms when a set of 5'- and 3'-doubly labeled probes (probes labeled with two different fluorochromes) is used. In addition, using dual-labeled probes is particularly relevant for environmental microorganisms since the fluorescence intensity signal is almost twice that of traditional FISH, without affecting the specificity.
- CLASI-FISH. In this approach, the species might be labeled with a combination of two (or more) versions of the sequencing probe, each version conjugated to a different fluorochrome. However, the probes compete by the same target site, which leads to a loss of fluorescence intensity signal; hence, cells with low ribosome content might not be correctly identified. This problem could be solved if two (or more) different probes labeled to a different fluorochrome, targeting different regions of rRNA, are used. However, in this approach, the number of probes is higher, and thus a hard probe design and selection might be done to ensure a set of probes that work in well-controlled experiments (e.g., same hybridization temperature).
FISH-Based Techniques for the Study of Metabolic Activity of Biofilm Cells
Biofilms exhibit dynamic processes that change according to a different stimulus; in addition, the phylogenetic identification does not provide any metabolic details of the identified microorganisms. In this sense, the FISH evolution has provided a possibility to identify, locate, and characterize complex communities to the functional level, with the development of some methods such as MicroAutoRadiography (MAR-FISH), Nanoscale secondary ion mass spectrometry (FISH-NanoSIMS) and Bio-orthogonal noncanonical amino acid tagging (BONCAT-FISH).
- MAR-FISH. MAR is a powerful tool, that allows the determination of the uptake of specific radioisotopes by individual microorganisms and has been used to study the in situ metabolic activity of microorganisms. This method is based on the assimilation of a radiolabeled substrate by individual cells, visualized by exposure to a radiation-sensitive silver halide emulsion placed on top of the radiolabeled bacteria and then processed by standard photographic techniques.
- FISH-NanoSIMS. NanoSIMS can be combined with FISH, in one technique called FISH-NanoSIMS, to directly link the cell identity and location to their activity. However, this analysis is not performed simultaneously; first, epifluorescence microscopy or a CLSM is used to identify and locate the microorganisms directly on samples. Then, the regions of interest will be analyzed by nanoSIMS instrument.
- BONCAT-FISH. This technique is based on the in vivo incorporation of the noncanonical amino acid L-azidohomoalanine (AHA), which is a surrogate for L-methionine, followed by fluorescent labeling of AHA-containing cellular proteins by azide-alkyne click chemistry. This technique when combined with FISH enables a link between in situ identification and translational activity in environmental biofilms at a single-cell level.
Creative Bioarray Relevant Recommendations
| Service/Product Types | Description |
| Fluorescent In Situ Hybridization (FISH) Service | Creative Bioarray offers a range of different FISH services including metaphase and interphase FISH, fibre-FISH, RNA-FISH, M-FISH, 3D-FISH, flow-FISH, FISH on paraffin sections, and immune-FISH. |
| Probe | Creative Bioarray provides the most comprehensive list of FISH probes for rapid identification of a wide range of chromosomal aberrations across the genome. |
| FISH Probe Design, Synthesis, and Testing Service | Creative Bioarray is capable of developing custom FISH probes. Apart from that, we also offer mRNA ISH/FISH probes, miRNA ISH/FISH probes, and lncRNA ISH/FISH probes. |
| FISH Quality Control Services | Creative Bioarray has years of experience in performing FISH, and cytogenetic services, we can also provide custom FISH quality control service for your lab's needs! |
RELATED PRODUCTS & SERVICES
Reference
- Barbosa A, et al. (2023). "Imaging biofilms using fluorescence in situ hybridization: seeing is believing." Front Cell Infect Microbiol. 13, 1195803.