Kasumi-3
Cat.No.: CSC-C6875J
Species: Homo sapiens (Human)
Source: Bone Marrow
Morphology: Myeloblast
- Specification
- Background
- Scientific Data
- Q & A
- Customer Review
The Kasumi-3 cell line is a unique and valuable resource in the field of leukemia research. This cell line was established in 1991 from the blast cells of a patient diagnosed with myeloperoxidase-negative acute leukemia. Myeloperoxidase (MPO) is an enzyme typically expressed in myeloid cells, and its absence is a characteristic feature of certain subtypes of acute leukemia. The Kasumi-3 cell line, derived from a patient with this rare MPO-negative acute leukemia, provides researchers with a relevant model system to study the underlying mechanisms and potential therapeutic approaches for this specific leukemic entity.
Extensive characterization of the Kasumi-3 cell line has revealed its phenotypic and genetic profile, which aligns with the patient's clinical presentation. The cells exhibit a range of surface markers and chromosomal abnormalities that are hallmarks of the specific leukemic subtype from which they were derived. The detailed understanding of the Kasumi-3 cell line's characteristics has made it an invaluable tool for researchers investigating the pathogenesis, signaling pathways, and potential therapeutic targets in MPO-negative acute leukemia.
Establishment of Kasumi-3 Cell Line With t(3;7)(q27;q22)
A novel human leukemia cell line (Kasumi-3) was established from the blast cells of a 57-year-old man suffering from myeloperoxidase-negative acute leukemia. The morphology of the cells is consistent with that of immature lymphoblasts, these cells being uniform in size with abundant basophilic cytoplasm, multiple nucleoli, and no granules (Fig. 1). The original leukemia cells and the Kasumi-3 cell line were positive for AP and negative for MPO, NBE, CAE, PAS, and NAP.
Both the original leukemia cells and the Kasumi-3 cells were positive for CD7, CD4, CD13, CD25, CD33, CD34, and HLA-DR. Surface and cytoplasmic CD3, CD36, CD41, and CD42 were negative. The Kasumi-3 cells and the original leukemic cells had common chromosomal abnormalities: 46, XY, t(3;7) (q27;q22), del(5)(q15), -8, del(9)(q32), add(12)(p11), +mar (Fig. 2). Additional abnormalities, t(2;5) (p13; q33) and add(16)(q13), appeared 10 months after establishment of the cell line. Southern blot hybridization showed no rearrangement bands with lgH, TCRβ, TCRγ, or TCRδ gene probes.
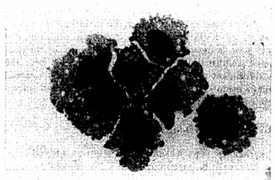 Fig. 1 Morphology of Kasumi-3 cells (MGG staining). (Asou H, et al., 1996)
Fig. 1 Morphology of Kasumi-3 cells (MGG staining). (Asou H, et al., 1996)
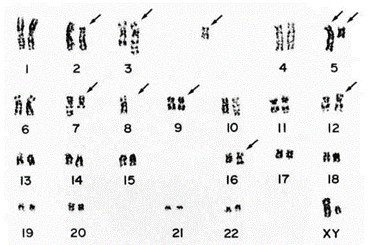 Fig. 2 Respresentative karyotype of Kasumi-3 cells. (Asou H, et al., 1996)
Fig. 2 Respresentative karyotype of Kasumi-3 cells. (Asou H, et al., 1996)
RNA Expression in Kasumi-3 Cells Infected by HCMV
Human cytomegalovirus (HCMV) establishes latency in myeloid cells. To investigate the kinetics of viral gene expression in Kasumi-3 cells at early times, genome-wide RNA expression at 4 and 24 h postinfection (hpi) was analyzed by transcriptome sequencing (RNA-seq) (Fig. 3A). In the Kasumi-3 model, 20% to 50% of the cells become infected, and the majority of the cells are therefore uninfected. To obtain a pure population of infected cells, additional studies were performed on cells that were sorted for GFP expression at 24 hpi (Fig. 3B).
In unsorted cells at 4 hpi, expression of the major immediate early genes UL122 and UL123 vastly exceeded that of other genes (Fig. 4). By 24 hpi, the pattern of viral RNA expression shifted significantly (Fig. 4 and 5), with a striking downregulation of the two major immediate early genes. Several other genes downstream of the major IE promoter (MIEP), such as the 5-kb intron RNA, UL119, UL120, UL121, and UL124 (Fig. 5) were downregulated, and there was a correlation between distance from the MIEP and the fold change in expression. In addition to the IE genes, UL16, UL23, UL37, UL91, UL100, UL116, UL134, UL137, US7, US33, and US34 were downregulated. Two early noncoding RNAs (ncRNAs), RNA2.7 and 1.2, were the most prominent transcripts upregulated at this time (Fig. 4). However, expression of the viral late gene transactivators UL79, UL87, UL91, UL92, and UL95 and the late genes, such as UL32, UL75, UL99, and UL115, was relatively low at this time. An exception was the late gene UL132, which was expressed at relatively high levels at 24 hpi in Kasumi-3 cells. Changes in the relative expression of selected viral genes from the immediate early (UL122 and UL123), early (UL54), and late (UL32) phases of viral replication were confirmed by reverse transcriptase quantitative PCR (RT-qPCR) (Fig. 4C).
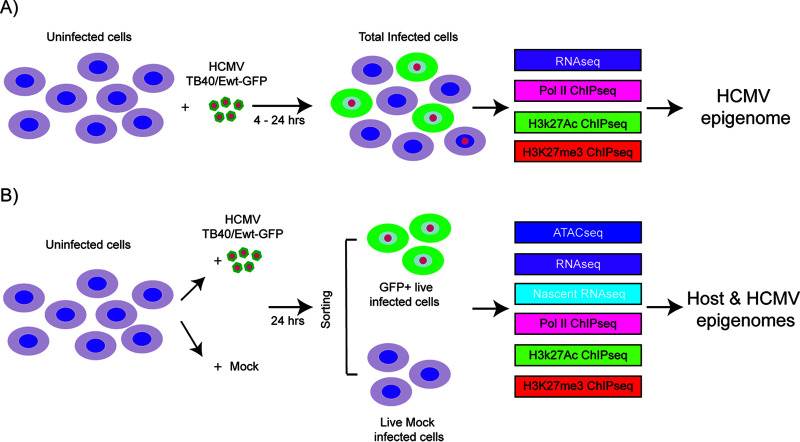 Fig. 3 Schematic of the samples used in the analysis of host and HCMV transcriptomes and epigenomes in Kasumi-3 cells. (Forte E, et al., 2021)
Fig. 3 Schematic of the samples used in the analysis of host and HCMV transcriptomes and epigenomes in Kasumi-3 cells. (Forte E, et al., 2021)
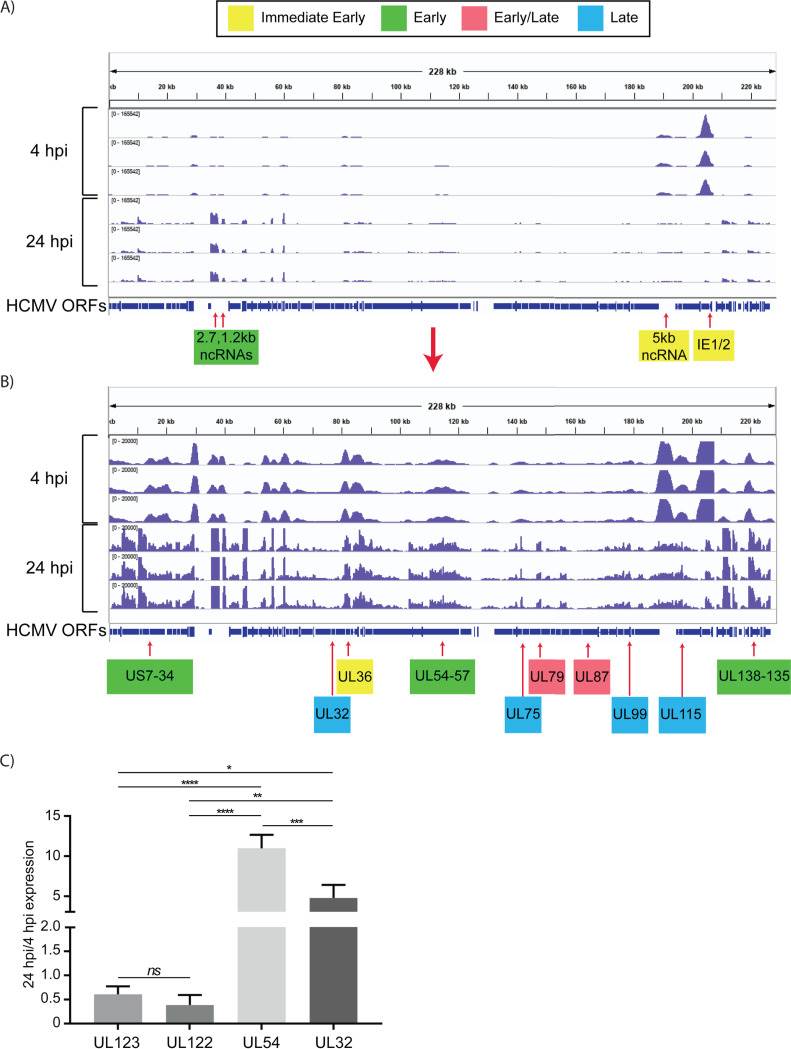 Fig. 4 Expression of HCMV RNAs in infected Kasumi-3 cells at 4 and 24 hpi. (Forte E, et al., 2021)
Fig. 4 Expression of HCMV RNAs in infected Kasumi-3 cells at 4 and 24 hpi. (Forte E, et al., 2021)
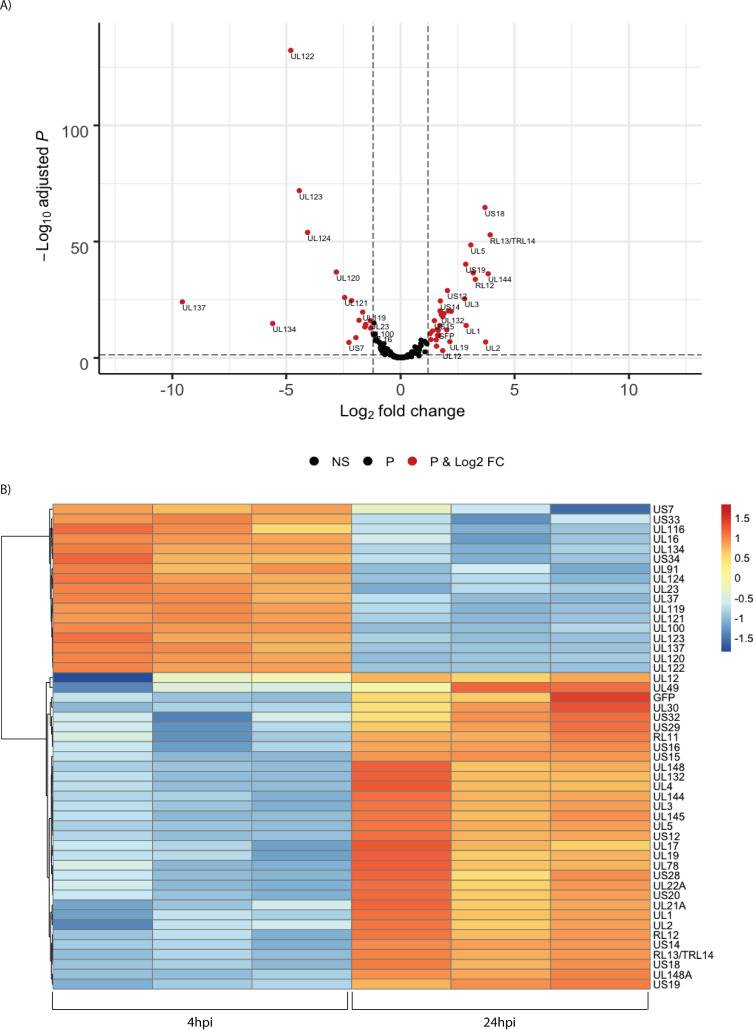 Fig. 5 Temporal changes in the relative abundance of HCMV RNAs. (Forte E, et al., 2021)
Fig. 5 Temporal changes in the relative abundance of HCMV RNAs. (Forte E, et al., 2021)
Ask a Question
Write your own review
- You May Also Need
- Adipose Tissue-Derived Stem Cells
- Human Neurons
- Mouse Probe
- Whole Chromosome Painting Probes
- Hepatic Cells
- Renal Cells
- In Vitro ADME Kits
- Tissue Microarray
- Tissue Blocks
- Tissue Sections
- FFPE Cell Pellet
- Probe
- Centromere Probes
- Telomere Probes
- Satellite Enumeration Probes
- Subtelomere Specific Probes
- Bacterial Probes
- ISH/FISH Probes
- Exosome Isolation Kit
- Human Adult Stem Cells
- Mouse Stem Cells
- iPSCs
- Mouse Embryonic Stem Cells
- iPSC Differentiation Kits
- Mesenchymal Stem Cells
- Immortalized Human Cells
- Immortalized Murine Cells
- Cell Immortalization Kit
- Adipose Cells
- Cardiac Cells
- Dermal Cells
- Epidermal Cells
- Peripheral Blood Mononuclear Cells
- Umbilical Cord Cells
- Monkey Primary Cells
- Mouse Primary Cells
- Breast Tumor Cells
- Colorectal Tumor Cells
- Esophageal Tumor Cells
- Lung Tumor Cells
- Leukemia/Lymphoma/Myeloma Cells
- Ovarian Tumor Cells
- Pancreatic Tumor Cells
- Mouse Tumor Cells