RS4;11
Cat.No.: CSC-C0021
Species: Homo sapiens (Human)
Source: Bone Marrow
Morphology: single, relatively small, round cells growing in suspension
- Specification
- Background
- Scientific Data
- Q & A
- Customer Review
- Documents
The RS4;11 cell line is a well-known and widely used cell line in cancer research, particularly in the study of acute lymphoblastic leukemia (ALL). These cells were originally derived from the bone marrow of a 32-year-old woman with ALL L2 during her first relapse. Researchers have utilized RS4;11 cells to investigate genetic abnormalities, signaling pathways, and mechanisms of drug resistance in leukemia cells.
One notable feature of the RS4;11 cell line is its Philadelphia chromosome-negative status, which distinguishes it from a subset of ALL cases characterized by the presence of the Philadelphia chromosome and the BCR-ABL1 fusion gene. This genetic characteristic of RS4;11 cells makes them particularly suitable for studying Philadelphia chromosome-negative ALL biology and exploring therapeutic strategies specific to this subgroup of patients.
Furthermore, the RS4;11 cell line is highly sensitive to chemotherapeutic agents commonly used in the treatment of ALL, such as vincristine and daunorubicin. This sensitivity allows researchers to evaluate the efficacy of potential anti-leukemic agents and assess their mechanisms of action using RS4;11 cells as a representative model.
The RS4;11 Cell Line as a Model for Leukaemia With t (4; 11) (q21; q23)
Hematological malignancies harboring rearrangements of the KMT2A gene represent a unique subtype of leukemia, with phenotypic clinical manifestations, a rapid and aggressive onset, and a generally poor prognosis. Chromosomal translocations involving KMT2A often cause the formation of oncogenic fusion genes, such as the most common translocation t (4;11) (q21; q23) producing the KMT2A-AFF1 chimera. This study aimed to confirm and review the cytogenetic and molecular features of the KMT2A-rearranged RS4;11 cell lines.
The presence of the t (4; 11) (q21; q23) was confirmed by M-FISH (Fig. 1), from which both der (4) and der (11) could be identified. The involvement of the KMT2A gene was further confirmed by FISH using a dual-color break-apart probe encompassing the KMT2A locus at 11q23.3. One normal KMT2A allele could be seen as a yellow fusion signal, whereas disruption of the KMT2A region would result in one red and one green signal present on the der (11) and der (4), respectively (Fig. 3).
Arm-specific probes showed the complete coverage of one chromosome 7 in red, corresponding to the long arm (Fig. 2C). For single locus probes, this was confirmed by the presence of two additional signals for 7q22 and 7q36 on one chromosome 7 (Fig. 2D). M-FISH (Fig. 1) and FISH using WCP8 (Fig. 2E) revealed two copies of chromosome 8 different in size and centromere position. FISH using a single locus DNA probe for 8q24.3 (RP11-195E4) was carried out to investigate a possible duplication of this locus. Signals specific to this region were visible on opposite sides of the centromere, suggesting the presence of an isochromosome 8q (Fig. 2F). The dual-color probe XL CDKN2A/2B highlighted the presence of both chromosome 9 centromeres. However, the lack of both signals for 9p21 revealed a homozygous deletion of that region (Fig. 2G, H).
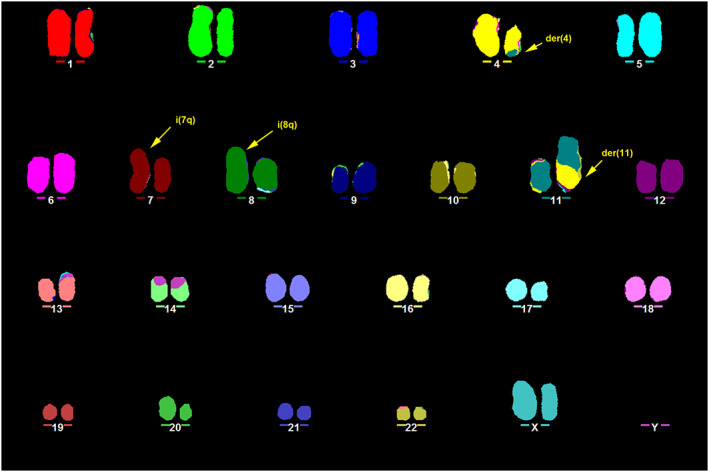 Fig. 1 Representative karyotype of RS4;11 obtained by M-FISH. (Ragusa D, et al., 2019)
Fig. 1 Representative karyotype of RS4;11 obtained by M-FISH. (Ragusa D, et al., 2019)
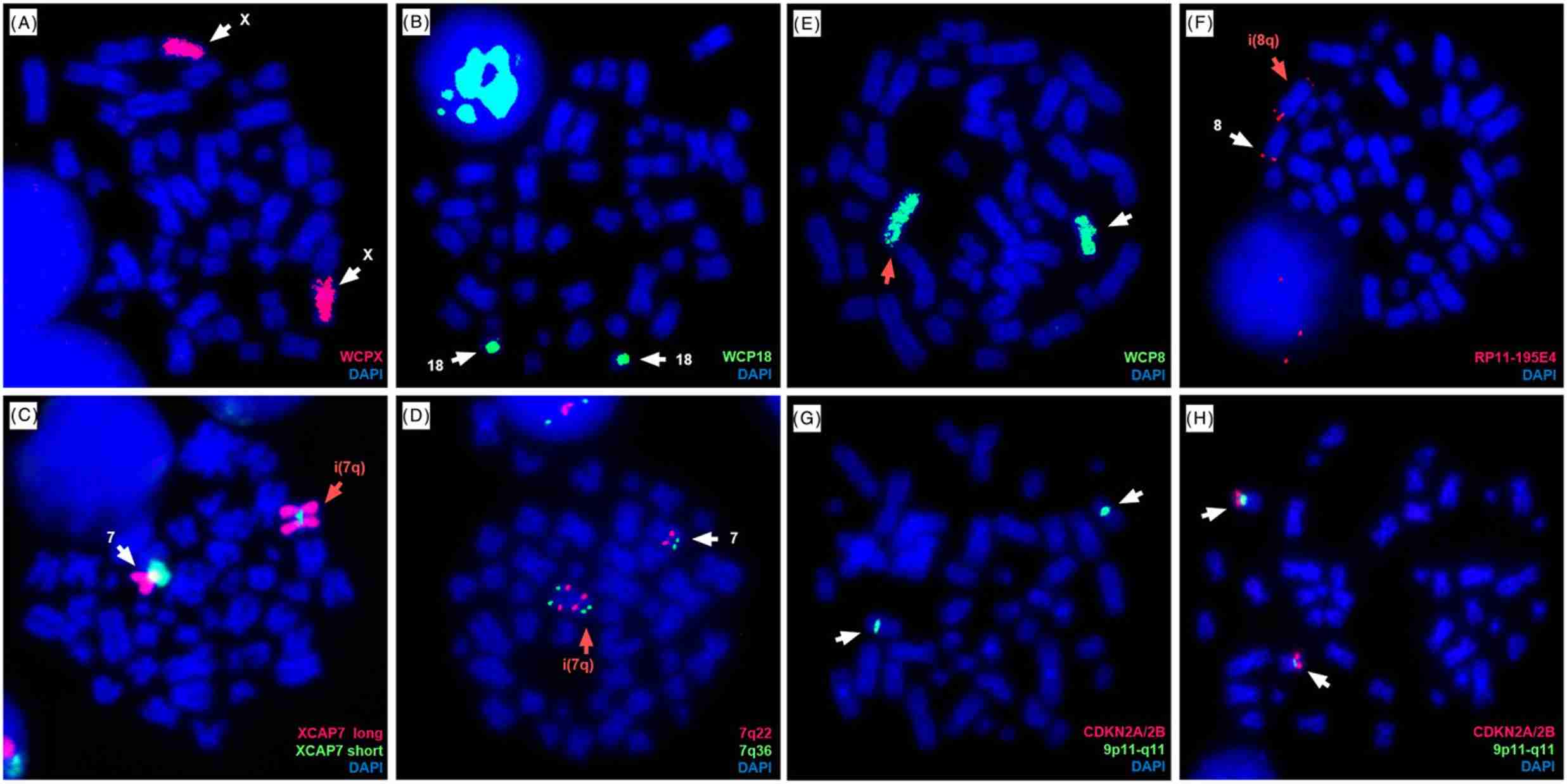 Fig. 2 Cytogenetic abnormalities investigated by FISH in the RS4;11 cell line. (Ragusa D, et al., 2019)
Fig. 2 Cytogenetic abnormalities investigated by FISH in the RS4;11 cell line. (Ragusa D, et al., 2019)
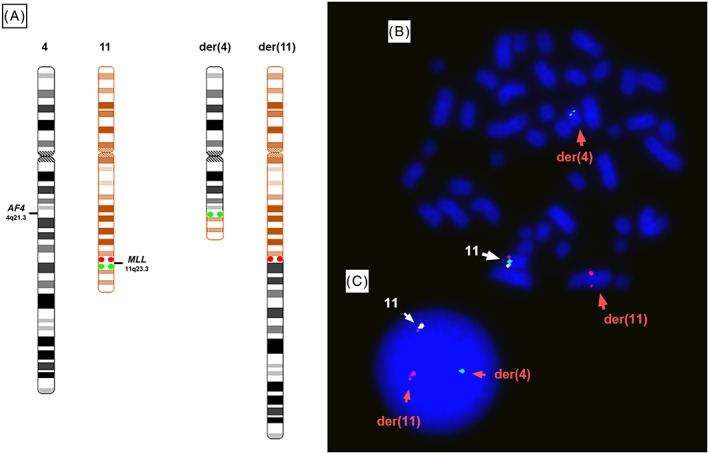 Fig. 3 Dual-colour FISH shows the KMT2A rearrangement in RS4;11. (Ragusa D, et al., 2019)
Fig. 3 Dual-colour FISH shows the KMT2A rearrangement in RS4;11. (Ragusa D, et al., 2019)
Differential Responses to SYK Inhibition in Precursor B-All Cells
The spleen tyrosine kinase (SYK) represents a potential therapeutic target in a subset of B-cell neoplasias. The biological and molecular effects of the SYK inhibitor entospletinib (Ento) in pre-B-ALL (NALM-6) and pro-B-ALL cell lines (SEM and RS4;11) were evaluated.
Ento exposure on pre-B-ALL NALM-6 (pre-BCR+) showed a concentration and slightly time-dependent decrease in cell proliferation. Similarly, Ento exposure also exhibited anti-proliferative effects on the pro-B-ALL cell line SEM (pre-BCR−). In contrast, the pro-B-ALL RS4;11 (pre-BCR−) showed only a moderate reduction in proliferation at the highest concentration of 10 µM and 20 µM applied for 48 h and 72 h (Fig. 4). High Ento concentrations induced significant early and late apoptosis/necrosis in the pre-B-ALL NALM-6 and SEM cell lines. In contrast, pro-B-ALL RS4;11 was almost resistant to apoptosis induction by Ento (Fig. 5). Cell-cycle distribution was investigated in three cell lines after Entospletinib exposure. Both concentrations of Ento (low concentration 1 µM or high concentration 10 µM) were not able to induce changes in cell-cycle distribution after 72 h incubation in the tested B-ALL cell lines (Fig. 6).
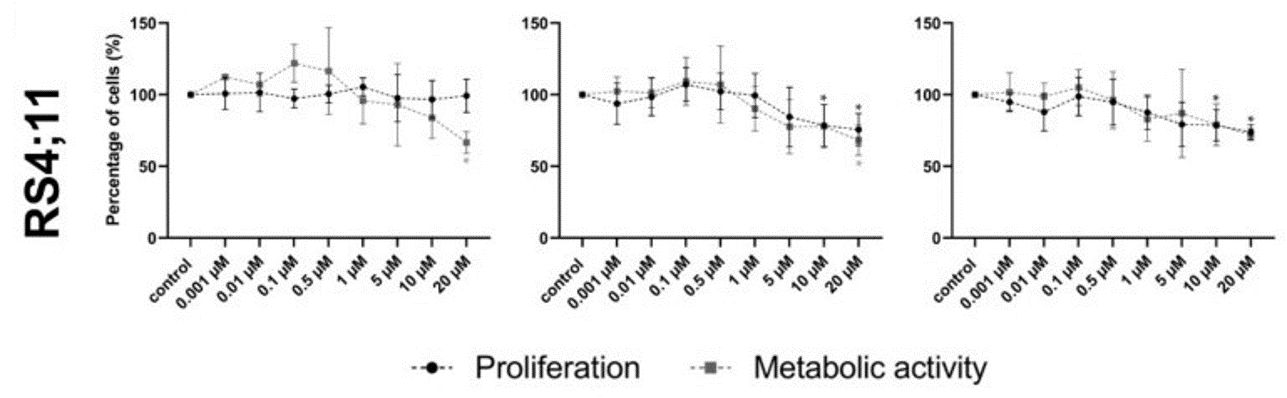 Fig. 4 Cell viability after entospletinib exposure in RS4;11 cell lines. (Sender S, et al., 2021)
Fig. 4 Cell viability after entospletinib exposure in RS4;11 cell lines. (Sender S, et al., 2021)
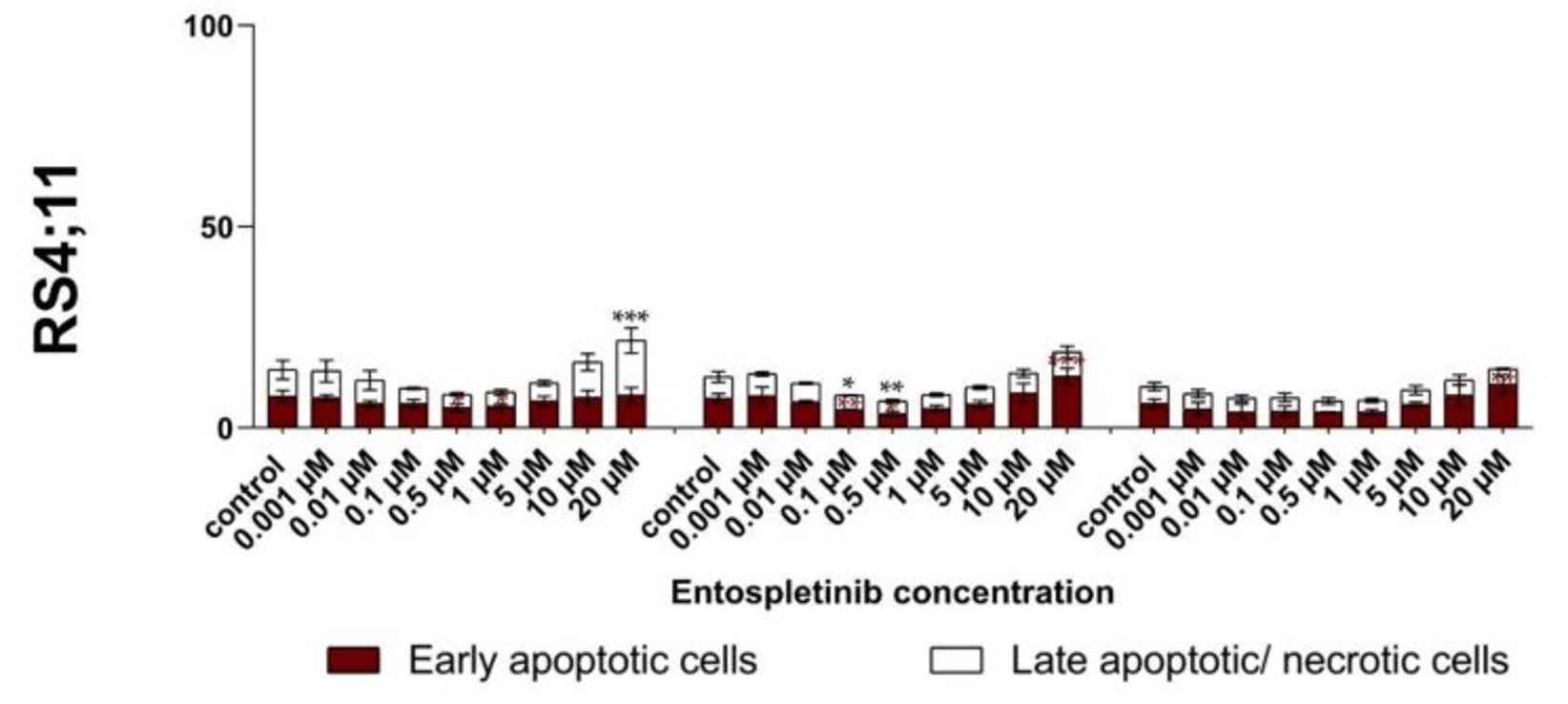 Fig. 5 Apoptosis induction in RS4;11 cells after entospletinib (Ento) exposure. (Sender S, et al., 221)
Fig. 5 Apoptosis induction in RS4;11 cells after entospletinib (Ento) exposure. (Sender S, et al., 221)
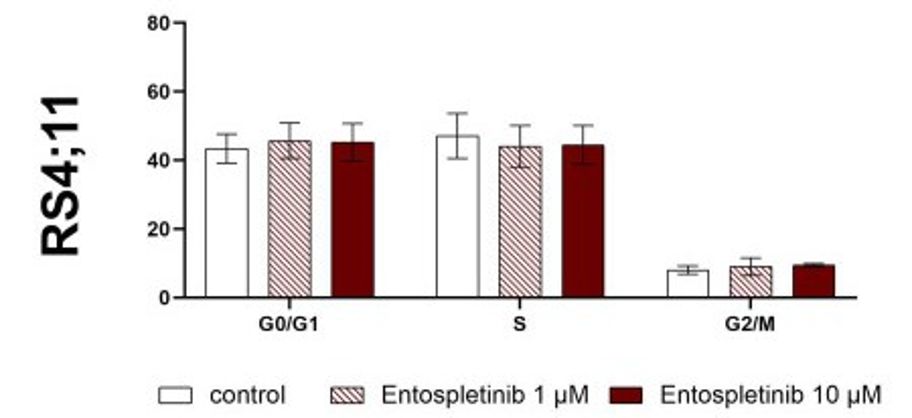 Fig. 6 Flow cytometric cell-cycle analysis of the RS4;11 cells after entospletinib exposure. (Sender S, et al., 2021)
Fig. 6 Flow cytometric cell-cycle analysis of the RS4;11 cells after entospletinib exposure. (Sender S, et al., 2021)
Ask a Question
Write your own review
- You May Also Need
- Adipose Tissue-Derived Stem Cells
- Human Neurons
- Mouse Probe
- Whole Chromosome Painting Probes
- Hepatic Cells
- Renal Cells
- In Vitro ADME Kits
- Tissue Microarray
- Tissue Blocks
- Tissue Sections
- FFPE Cell Pellet
- Probe
- Centromere Probes
- Telomere Probes
- Satellite Enumeration Probes
- Subtelomere Specific Probes
- Bacterial Probes
- ISH/FISH Probes
- Exosome Isolation Kit
- Human Adult Stem Cells
- Mouse Stem Cells
- iPSCs
- Mouse Embryonic Stem Cells
- iPSC Differentiation Kits
- Mesenchymal Stem Cells
- Immortalized Human Cells
- Immortalized Murine Cells
- Cell Immortalization Kit
- Adipose Cells
- Cardiac Cells
- Dermal Cells
- Epidermal Cells
- Peripheral Blood Mononuclear Cells
- Umbilical Cord Cells
- Monkey Primary Cells
- Mouse Primary Cells
- Breast Tumor Cells
- Colorectal Tumor Cells
- Esophageal Tumor Cells
- Lung Tumor Cells
- Leukemia/Lymphoma/Myeloma Cells
- Ovarian Tumor Cells
- Pancreatic Tumor Cells
- Mouse Tumor Cells