L-1236
Cat.No.: CSC-C0538
Species: Homo sapiens (Human)
Source: Blood; Peripheral Blood
Morphology: variable morphology with semi-adherent spheroid to fusiform cells growing in clumps in mono- and multilayers, occasional giant cells
- Specification
- Background
- Scientific Data
- Q & A
- Customer Review
Immunology: CD2 -, CD3 -, CD13 -, CD14 -, CD15 +, CD19 -, CD21 -, CD25 -, CD30 -, CD34 -, CD54 +, CD80 +
Viruses: PCR: EBV -, HBV -, HCV -, HIV -, HTLV-I/II -, SMRV -
The L-1236 cell line was established from the peripheral blood of a 34-year-old man diagnosed with Hodgkin lymphoma in 1994. The patient was specifically diagnosed with mixed cellularity subtype, stage IV, refractory, terminal, and third relapse, indicating an aggressive and advanced form of the disease. The establishment of the L-1236 cell line represents a critical development in cancer research, providing researchers with a valuable tool for studying the mechanisms of Hodgkin lymphoma and developing potential treatments.
Due to the aggressive nature of the patient's condition, the L-1236 cell line is of significant interest to researchers focusing on refractory and relapsed Hodgkin lymphoma. Over the years, the L-1236 cell line has been extensively utilized in various research studies aimed at understanding the underlying biology of Hodgkin lymphoma, including investigating potential therapeutic targets, unraveling drug resistance mechanisms, and exploring new treatment options. Its establishment has therefore played a crucial role in advancing our knowledge of Hodgkin lymphoma and has contributed to the ongoing efforts to improve patient outcomes through targeted therapies and personalized medicine approaches.
Amplification of Rearranged V Region Genes From Cell Line L1236
To analyze cell line L1236 for Ig gene rearrangements, three sets of oligonucleotides were used. A VH1 gene rearrangement was obtained with the VH1 leader primer, but not with the FRI primer. Sequence analysis of the PCR product revealed a potentially functional rearrangement of a VH1 gene with the highest homology to the HV1F10 VH1 germline gene (Fig. 1, L1236H1). There are 55 nucleotide differences between the VH gene segment of L1236H1 and the HV1F10 gene (i.e., 87% homology). A JH4 gene was used in the rearrangement, but no DH gene could be identified (Fig. 1).
A VH3 gene rearrangement of unexpected length was amplified with both the VH3 FRI and leader primer. The V gene segment was rearranged into the intron between JH3 and JH4, and there was a deletion of codons 30 to 33 in the complementarity-determining region (CDR) I and a duplication of codons 66 to 73 in FRIII (Fig. 2, L1236H3). Interestingly, close to the rearrangement breakpoint in the JH intron, a tetranucleotide sequence motif (5'CTGG375'CCAG3') commonly found near nonhomologous recombinations involving Ig gene sequences was found twice (Fig. 2). The same motif was also seen near the deletion in CDRI and within the duplication in FRIII (Fig. 2). A potentially functional VK3 gene rearrangement amplified from the genomic DNA of L1236 was composed of the L2 VK3 germline gene rearranged to JK1 (Fig. 3, L1236K3). The VK gene segment of L1236K3 carried 41 nucleotide differences versus L2 (84% homology). One mutation was found in the JKl gene segment (Fig. 3).
 Fig. 1 VH1 gene rearrangements from cell line L1236 and single Hodgkin and Reed-Sternberg (H-RS) cells. (Kanzler H, et al., 1996)
Fig. 1 VH1 gene rearrangements from cell line L1236 and single Hodgkin and Reed-Sternberg (H-RS) cells. (Kanzler H, et al., 1996)
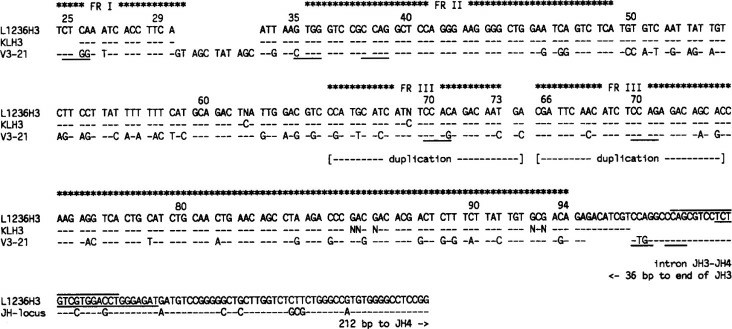 Fig. 2 VH3 gene rearrangements from cell line L1236 and single H-RS cells. (Kanzler H, et al., 1996)
Fig. 2 VH3 gene rearrangements from cell line L1236 and single H-RS cells. (Kanzler H, et al., 1996)
 Fig. 3 VK3 gene rearrangements derived from cell line L1236 and single H-RS cells. (Kanzler H, et al., 1996)
Fig. 3 VK3 gene rearrangements derived from cell line L1236 and single H-RS cells. (Kanzler H, et al., 1996)
Comparison of SAGE Profiles from the cHL Cell Line L1236 and GC B Cells
Serial analysis of gene expression (SAGE) profiles were generated from the cHL cell line L1236 and human GC B cells. For the isolation of GC B cells, CD77+ GC cells are used because gene expression analysis based on oligonucleotide arrays showed surprisingly few differences between gene expression patterns of tonsillar centrocytes (defined as CD10+ CD77−) and tonsillar centroblasts (defined as CD77+) (Fig. 4). When compared with GC B cells, the cell line L1236 showed 177 transcripts with higher expression and 287 with lower expression.
Analyzing the downregulated genes, a dramatic loss of B lineage-specific gene expression became obvious, affecting nearly all B-cell-specific genes. Only some genes involved in MHC class II-restricted antigen presentation are still expressed or even show an elevated expression, for example, CD40, B7.1, B7.2, and some MHC class II alleles. Our further studies concentrated on transcripts showing an increased expression in the cHL cell line L1236. Several transcripts known to be involved in or associated with malignant transformation are upregulated or specifically expressed in the L1236 cell line. This includes potential oncogenes, such as PTP4A, rhoC, or L-myc, cathepsins known to be important in tumor progression, and genes involved in cell adhesion, such as the laminin receptor or CD44. In addition to molecules playing a role in signaling cascades, such as Dvl-1 or Nm23-H1, several transcription factors show elevated expression.
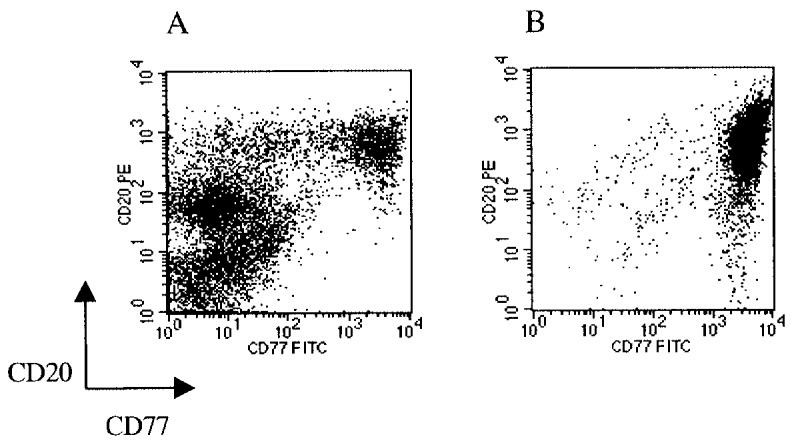 Fig. 4 FACS analysis of human tonsillar mononucleated cells after Ficoll purification (A) and after MACS enrichment of CD77+ cells (B). (Schwering I, et al., 2003)
Fig. 4 FACS analysis of human tonsillar mononucleated cells after Ficoll purification (A) and after MACS enrichment of CD77+ cells (B). (Schwering I, et al., 2003)
Ask a Question
Write your own review
- You May Also Need
- Adipose Tissue-Derived Stem Cells
- Human Neurons
- Mouse Probe
- Whole Chromosome Painting Probes
- Hepatic Cells
- Renal Cells
- In Vitro ADME Kits
- Tissue Microarray
- Tissue Blocks
- Tissue Sections
- FFPE Cell Pellet
- Probe
- Centromere Probes
- Telomere Probes
- Satellite Enumeration Probes
- Subtelomere Specific Probes
- Bacterial Probes
- ISH/FISH Probes
- Exosome Isolation Kit
- Human Adult Stem Cells
- Mouse Stem Cells
- iPSCs
- Mouse Embryonic Stem Cells
- iPSC Differentiation Kits
- Mesenchymal Stem Cells
- Immortalized Human Cells
- Immortalized Murine Cells
- Cell Immortalization Kit
- Adipose Cells
- Cardiac Cells
- Dermal Cells
- Epidermal Cells
- Peripheral Blood Mononuclear Cells
- Umbilical Cord Cells
- Monkey Primary Cells
- Mouse Primary Cells
- Breast Tumor Cells
- Colorectal Tumor Cells
- Esophageal Tumor Cells
- Lung Tumor Cells
- Leukemia/Lymphoma/Myeloma Cells
- Ovarian Tumor Cells
- Pancreatic Tumor Cells
- Mouse Tumor Cells