Pfeiffer
Cat.No.: CSC-C9234W
Species: Homo sapiens (Human)
Source: Pleural Effusion
Morphology: lymphoblast
- Specification
- Background
- Scientific Data
- Q & A
- Customer Review
The Pfeiffer cell line is a human B-cell lymphoma line which was first developed in the peripheral blood of a patient with diffuse large B-cell lymphoma (DLBCL). It is a mature B-cell line and used as a model for studying the biology and therapeutic targeting of aggressive lymphomas.
Functionally, Pfeiffer cells are representative of activated B-cells and possess several hallmark features of DLBCL. For example, Pfeiffer cells harbor the chromosomal translocation t(8;14), which results in the overexpression of the MYC oncogene. MYC overexpression in Pfeiffer cells leads to uncontrolled cell proliferation and metabolic reprogramming that is required for oncogenesis. This makes Pfeiffer cells an important tool to study MYC-driven cancers. Pfeiffer cells also have constitutive activation of the NF-κB pathway, which is a key feature required for their proliferation, survival, and resistance to apoptosis.
The Pfeiffer cell line is mainly used as a cancer research tool to study lymphomagenesis and drug resistance, as well as for preclinical testing of new drugs. Pfeiffer cells are a common cell line used to screen and validate the efficacy of small molecule inhibitors, particularly those targeting the B-cell receptor, NF-κB, and MYC. They are also a standard model to test the mechanism of action of chemotherapeutics and to develop immunotherapies including antibody-drug conjugates. Their high proliferative capacity and genetic tractability also make these cells a useful tool for in vitro and in vivo (xenograft) experiments to understand tumor biology and test combination therapies.
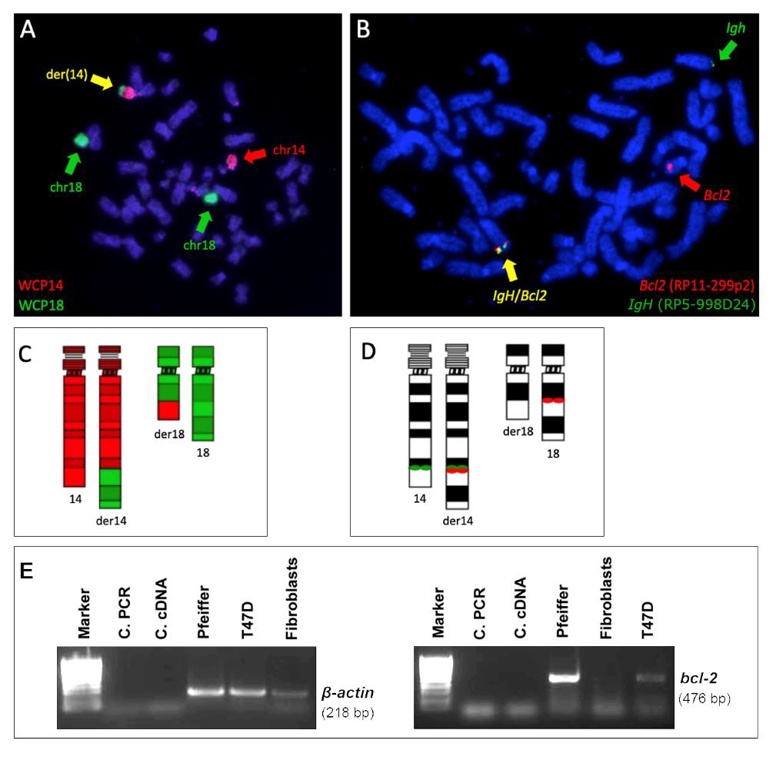
t(14;18) Translocation and BCL2 Expression in the Pfeiffer Cell Line
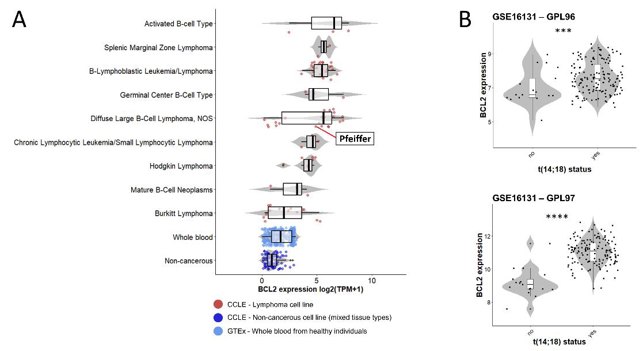
Chromosomal rearrangements can alter genome organization and gene expression. In leukemia, nuclear positioning of fusion genes from chromosomal translocations can exacerbate gene expression, but this is understudied in lymphoma. Garimberti et al. explored genome organization in lymphoma cells Pfeiffer with the t(14;18)(q32;q21) rearrangement, which overexpresses the BCL2 gene, using fluorescence in situ hybridization (FISH) to map chromosome territories and translocated genes in the Pfeiffer cell line.
The t(14;18) translocation in the Pfeiffer cell line was confirmed by FISH. Due to the limited extent of the translocated portion from chromosome 14, der(18) was undetectable with whole chromosome painting (WCP) (Fig. 1A). However, using both WCP and bacterial artificial clone (BAC) probes, they clearly identified der(14) and the IGH::BCL2 gene fusion (Fig. 1). This translocation correlates with BCL2 expression in the Pfeiffer cell line and in lymphoma patients (Fig. 2). RNA-seq data from the Cancer Cell Line Encyclopedia (CCLE) showed high BCL2 expression in diffuse large B-cell lymphoma lines, including Pfeiffer, and low expression in whole blood and non-cancerous tissue (Fig. 2A). BCL2 expression in Pfeiffer cells was also confirmed by in-house reverse transcription (RT) PCR (Fig. 1E). Additionally, BCL2 expression was significantly higher in follicular lymphoma patients with t(14;18) compared to those without, based on publicly available data (GSE16131) (Fig. 2B).
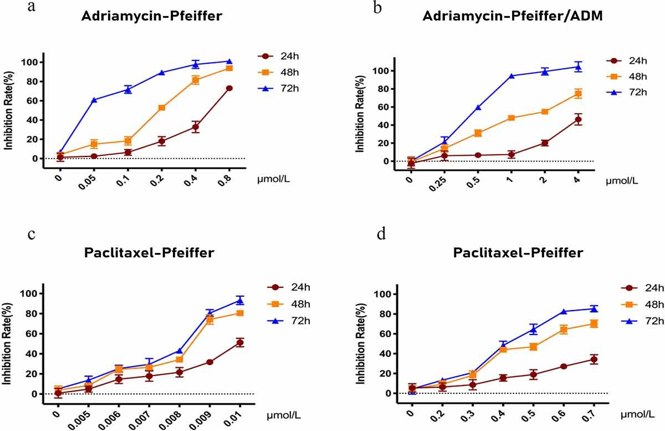
RNA-seq Analysis Reveals the Hub Genes of Paclitaxel Inhibition on Adriamycin Resistant DLBCL Cells
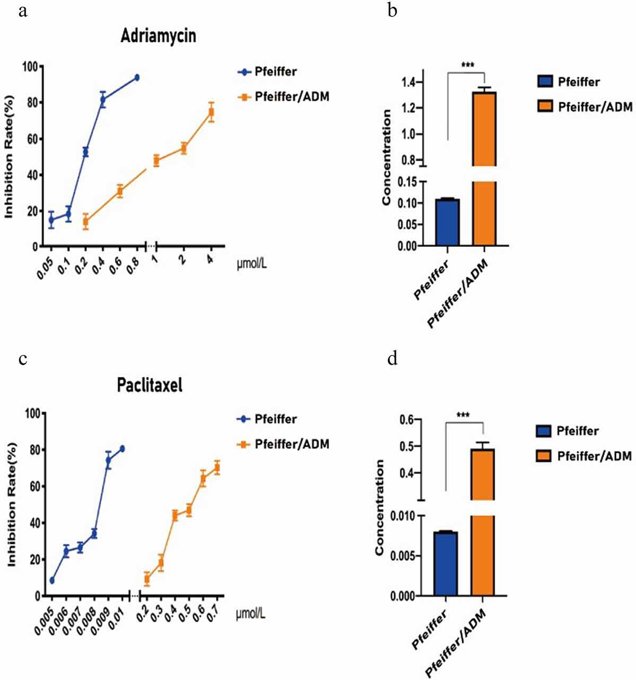
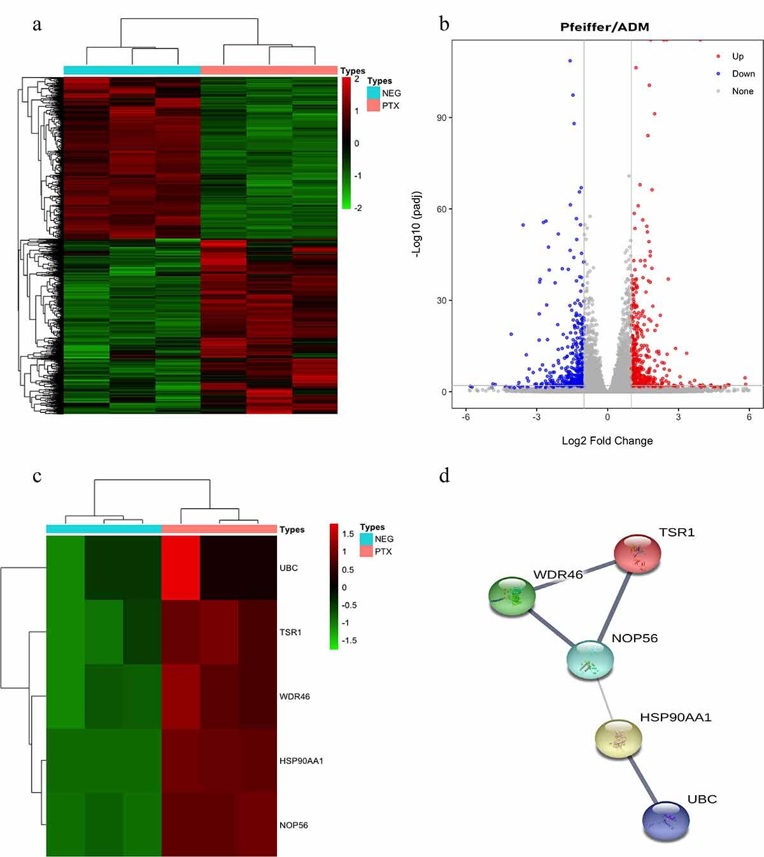
40% of patients with diffuse large B-cell lymphoma (DLBCL) are insensitive to first-line chemotherapy, in which Adriamycin is the main drug. The previous work showed that Paclitaxel may be a therapeutic for Adriamycin-resistant DLBCL. Here, Hong et al. sought to elucidate the molecular mechanisms by which Paclitaxel treats Adriamycin-resistant DLBCL using RNA-seq and network analysis. CCK-8 assay results showed that Paclitaxel and Adriamycin significantly inhibited Pfeiffer and Pfeiffer/ADM cell proliferation in a dose- and time-dependent manner (Fig. 3). The IC50 of Paclitaxel in Pfeiffer/ADM cells was 0.4912 ± 0.0230 μmol/L, significantly lower than Adriamycin (1.3256 ± 0.0328 μmol/L), indicating that the sensitivity of Paclitaxel was higher (Fig. 4). RNA sequencing results identified 971 differentially expressed genes (DEGs), which included 519 upregulated and 452 downregulated genes (Fig. 5).
Ask a Question
Write your own review
- You May Also Need
- Adipose Tissue-Derived Stem Cells
- Human Neurons
- Mouse Probe
- Whole Chromosome Painting Probes
- Hepatic Cells
- Renal Cells
- In Vitro ADME Kits
- Tissue Microarray
- Tissue Blocks
- Tissue Sections
- FFPE Cell Pellet
- Probe
- Centromere Probes
- Telomere Probes
- Satellite Enumeration Probes
- Subtelomere Specific Probes
- Bacterial Probes
- ISH/FISH Probes
- Exosome Isolation Kit
- Human Adult Stem Cells
- Mouse Stem Cells
- iPSCs
- Mouse Embryonic Stem Cells
- iPSC Differentiation Kits
- Mesenchymal Stem Cells
- Immortalized Human Cells
- Immortalized Murine Cells
- Cell Immortalization Kit
- Adipose Cells
- Cardiac Cells
- Dermal Cells
- Epidermal Cells
- Peripheral Blood Mononuclear Cells
- Umbilical Cord Cells
- Monkey Primary Cells
- Mouse Primary Cells
- Breast Tumor Cells
- Colorectal Tumor Cells
- Esophageal Tumor Cells
- Lung Tumor Cells
- Leukemia/Lymphoma/Myeloma Cells
- Ovarian Tumor Cells
- Pancreatic Tumor Cells
- Mouse Tumor Cells