MEL-745A cl. DS19
Cat.No.: CSC-C3447
Species: Mus musculus (Mouse)
Morphology: round, single or in small clusters, growing in suspension, about 5% giant cells
Culture Properties: suspension
- Specification
- Background
- Scientific Data
- Q & A
- Customer Review
Cytogenetics: murine hypertetraploid karyotype
Viruses: PCR: SMRV -
The MEL-745A cl. DS19 cell line developed from mouse erythroleukemia cells which transformed under the influence of the Friend virus retrovirus. The DS19 cell line responds to dimethyl sulfoxide (DMSO) by differentiating into mature red blood cells through DMSO induction. This differentiation process is accompanied by increased hemoglobin synthesis and a reduction in malignant phenotype. Research indicates that during differentiation this cell line shows control over particular gene expression patterns including the activation of β-globin gene transcription. Additionally, this cell line responds to chemical inducers such as HMBA by undergoing differentiation to reach the terminal differentiation stage.
MEL-745A cl. DS19 cell line serves as a common research model for scientists studying blood disorders and cancer. For example, researchers use CRISPR-Cas9 technology to target and edit genes like the BCL11A enhancer to analyze their impact on erythrocyte differentiation.
Stable Transfection of MEL Cells with Anti-Gpx4 Antisense Construct Reduced Gpx4 Expression
Glutathione peroxidase 4 (GPX4), unique in reducing lipid peroxides, is vital for embryonic development and physiological functions. Previous studies showed Gpx4 knockout causes embryonic lethality in mice, and conditional silencing in erythroid precursors impaired erythropoiesis. However, the role of GPX4 in specific in vitro erythroid differentiation remained unclear. Rademacher et al. investigated GPX4's role in erythropoiesis using mouse erythroleukemia cells (MEL-745A cl. DS19).
To determine if MEL cells express Gpx4 isoforms, they performed qRT-PCR to measure Gpx4 mRNA levels in resting cells (Fig. 1a). The cytosolic isoform, cGpx4, showed high expression, with over 150 mRNA copies per 1000 Gapdh mRNA copies. The mitochondrial isoform, mGpx4, had four times lower expression, and nuclear Gpx4 mRNA was less than 1% of cGpx4 levels. This expression pattern aligns with other mouse tissues. To assess the effect of Gpx4 silencing, they used siRNA technology to create a model by stably transfecting MEL cells with an anti-Gpx4 plasmid and analyzing surviving clones. Three distinct cell clones were selected. All exhibited about 50% Gpx4 expression silencing (Fig. 1b). Gapdh expression remained constant, confirming unaffected reference gene expression (Fig. 1b inset). Clone 2, with 51 ± 2% Gpx4 silencing, was chosen for detailed studies.
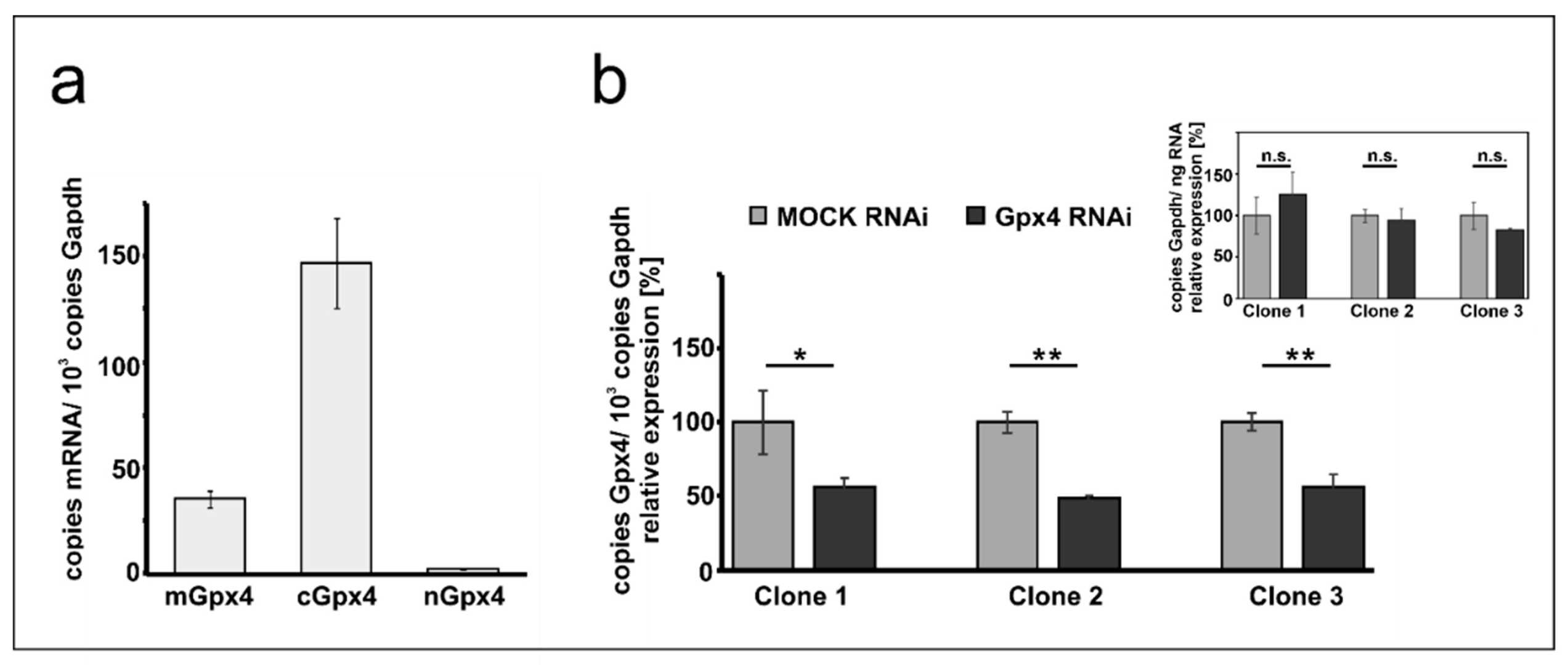 Fig. 1. Expression of Gpx4 isoforms in native and transfected MEL cells (Rademacher M, Kuhn H, et al., 2021).
Fig. 1. Expression of Gpx4 isoforms in native and transfected MEL cells (Rademacher M, Kuhn H, et al., 2021).
The Role of Crowding Forces in Juxtaposing β-Globin Gene Domain Remote Regulatory Elements in Mouse Erythroid Cells
The eukaryotic cell nucleus is crowded with macromolecules, leading to crowding forces that help maintain nuclear compartment integrity. While these forces are known to influence nuclear structures, their role in chromatin dynamics is not well-understood. Golov et al. investigated this role by examining the spatial organization of the β-globin domain in mouse erythroid cells.
In all experiments, they used murine adult erythroleukemia cell line MEL (MEL-745A cl. DS19) to study the β-globin ACH configuration. They modified the protocol for nuclei isolation and expansion to suit MEL cells, following Hancock's method. MEL cells, lysed in isotonic solution with digitonin, retained nuclear integrity, while small molecules exited through nuclear pores. Proteins such as GATA–1, DNA topoisomerase I, and PML were tested for retention in nuclei after treatments. We found ~90% GATA–1 retention, while DNA topoisomerase I and PML were almost fully retained (Fig. 2). This suggests that macromolecular crowding levels in isolated nuclei are similar to intact cells. Expanding nuclei in hypotonic buffer reduced crowding due to redistribution of macromolecules and loss of small proteins like GATA–1.
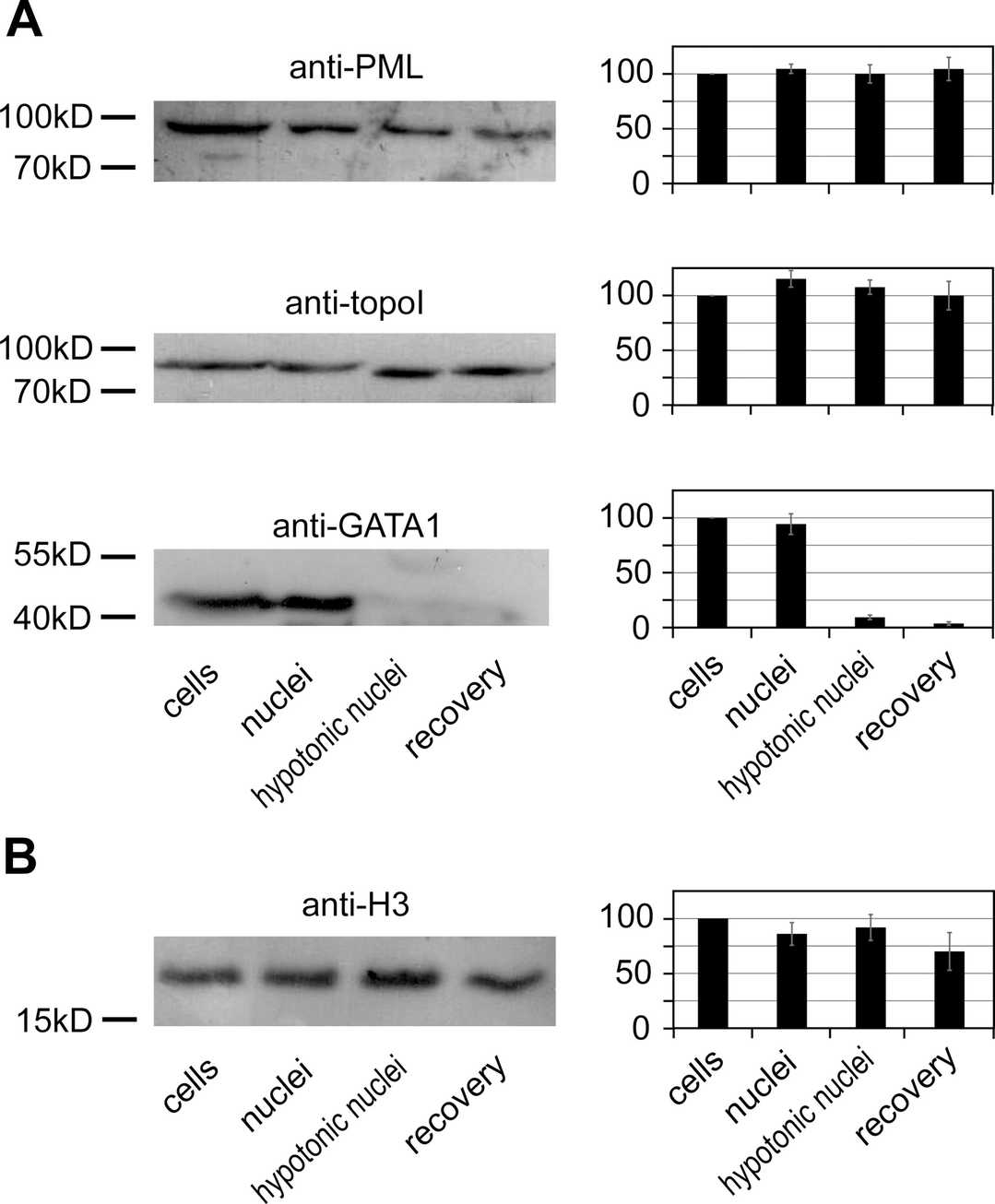 Fig. 2. Retention of PML, DNA topoisomerase I, GATA–1, and histone H3 in nuclei after cell lysis and subsequent treatments (Golov A K, Gavrilov A A, et al., 2015).
Fig. 2. Retention of PML, DNA topoisomerase I, GATA–1, and histone H3 in nuclei after cell lysis and subsequent treatments (Golov A K, Gavrilov A A, et al., 2015).
Ask a Question
Write your own review
- You May Also Need
- Adipose Tissue-Derived Stem Cells
- Human Neurons
- Mouse Probe
- Whole Chromosome Painting Probes
- Hepatic Cells
- Renal Cells
- In Vitro ADME Kits
- Tissue Microarray
- Tissue Blocks
- Tissue Sections
- FFPE Cell Pellet
- Probe
- Centromere Probes
- Telomere Probes
- Satellite Enumeration Probes
- Subtelomere Specific Probes
- Bacterial Probes
- ISH/FISH Probes
- Exosome Isolation Kit
- Human Adult Stem Cells
- Mouse Stem Cells
- iPSCs
- Mouse Embryonic Stem Cells
- iPSC Differentiation Kits
- Mesenchymal Stem Cells
- Immortalized Human Cells
- Immortalized Murine Cells
- Cell Immortalization Kit
- Adipose Cells
- Cardiac Cells
- Dermal Cells
- Epidermal Cells
- Peripheral Blood Mononuclear Cells
- Umbilical Cord Cells
- Monkey Primary Cells
- Mouse Primary Cells
- Breast Tumor Cells
- Colorectal Tumor Cells
- Esophageal Tumor Cells
- Lung Tumor Cells
- Leukemia/Lymphoma/Myeloma Cells
- Ovarian Tumor Cells
- Pancreatic Tumor Cells
- Mouse Tumor Cells