KYSE-150
Cat.No.: CSC-C0423
Species: Homo sapiens (Human)
Source: Esophagus
Morphology: epitheloid cells growing as adherent monolayer
Culture Properties: monolayer
- Specification
- Background
- Scientific Data
- Q & A
- Customer Review
Immunology: cytokeratin +, cytokeratin-7 -, cytokeratin-8 +, cytokeratin-17 +, cytokeratin-18 +, cy
The KYSE-150 cell line is a human esophageal squamous cell carcinoma (ESCC) model derived from upper esophagus of a 49-year-old woman after receiving radiotherapy. This cell line is part of the KYSE series designed to provide a reliable in vitro model for studying the pathobiology of esophageal cancer, particularly in understanding tumorigenesis and therapeutic response.
KYSE-150 cells exhibit a rapid doubling time of 13.7 hours, indicating a high proliferative capacity, which is characteristic of an aggressive cancer phenotype. These cells grow in monolayer culture, adhering to substrates and form uniform sheets, which is typical for epithelial-derived cancer cells. Genetic analysis of KYSE-150 reveals significant alterations in key tumor suppressor genes, particularly the p16 (INK4a) gene. The cell line shows p16 gene aberrations, specifically in the form of CpG island methylation, silencing this gene and leading to loss of cell cycle regulation. This epigenetic modification is a common mechanism in many cancers and highlights the relevance of KYSE-150 for studying gene silencing and its role in cancer progression. Furthermore, the cell line retains the wild-type configuration of the p15 gene, suggesting a selective inactivation mechanism for p16 over p15 in this model, which may be of interest in comparative genomic studies.
KYSE-150 is valuable not only for studying the molecular and cellular mechanisms of ESCC, but also for exploring the role of genetic and epigenetic alterations in cancer. It provides a powerful model for investigating therapeutic interventions targeting specific pathways dysregulated in ESCC. Given its high proliferation rate and specific genetic profile, KYSE-150 is a suitable candidate for in vitro pharmacological testing and other applications related to cancer research.
The Genomic Landscape of Esophageal Squamous Cell Carcinoma (ESCC) Cell Lines
Whole exome and RNA sequencing were conducted on ESCC cell lines TE-1, ECA-109, KYSE-30, KYSE-150, KYSE-180, KYSE-450, and KYSE-510, with the normal epithelium cell line Het-1a as a comparison. Bioinformatics methods analyzed gene mutation types, mutation frequencies, RNA expression, and classical cancer-related signaling pathways. Specific analyses were also performed on tumor burden, genes related to differentiation, invasion, immunity, and gene enrichment in each cell line.
This study aims to elucidate the malignancy, invasion capability, classical cancer-related signaling pathways, and immune status of ESCC cell lines, providing a detailed genomic landscape and highlighting the unique features of each cell line. This genomic landscape offers valuable insights for future research and therapeutic strategies in ESCC.
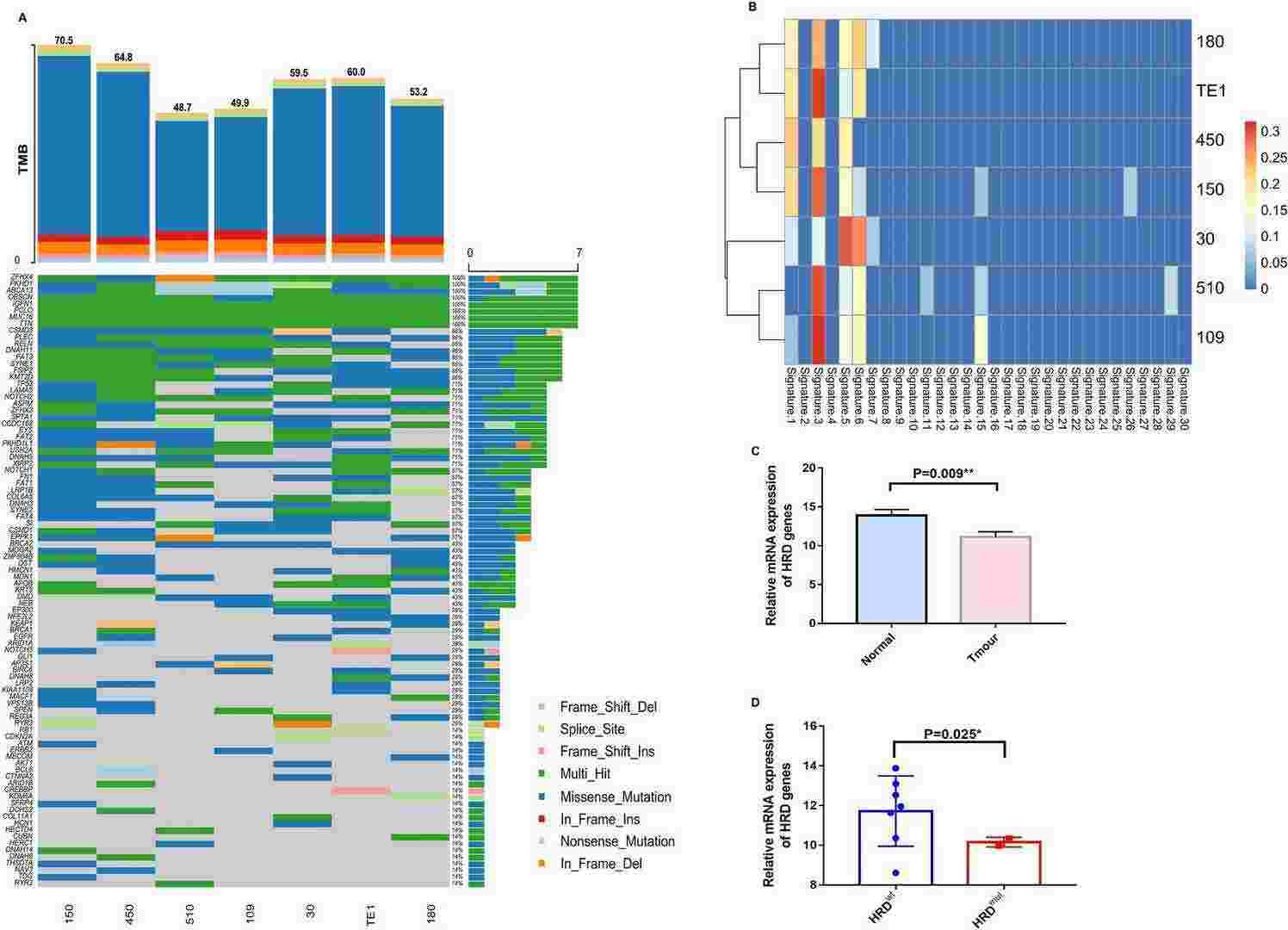 Fig. 1. Mutational signatures of ESCC cell lines (Zhang, Chao, et al. 2025).
Fig. 1. Mutational signatures of ESCC cell lines (Zhang, Chao, et al. 2025).
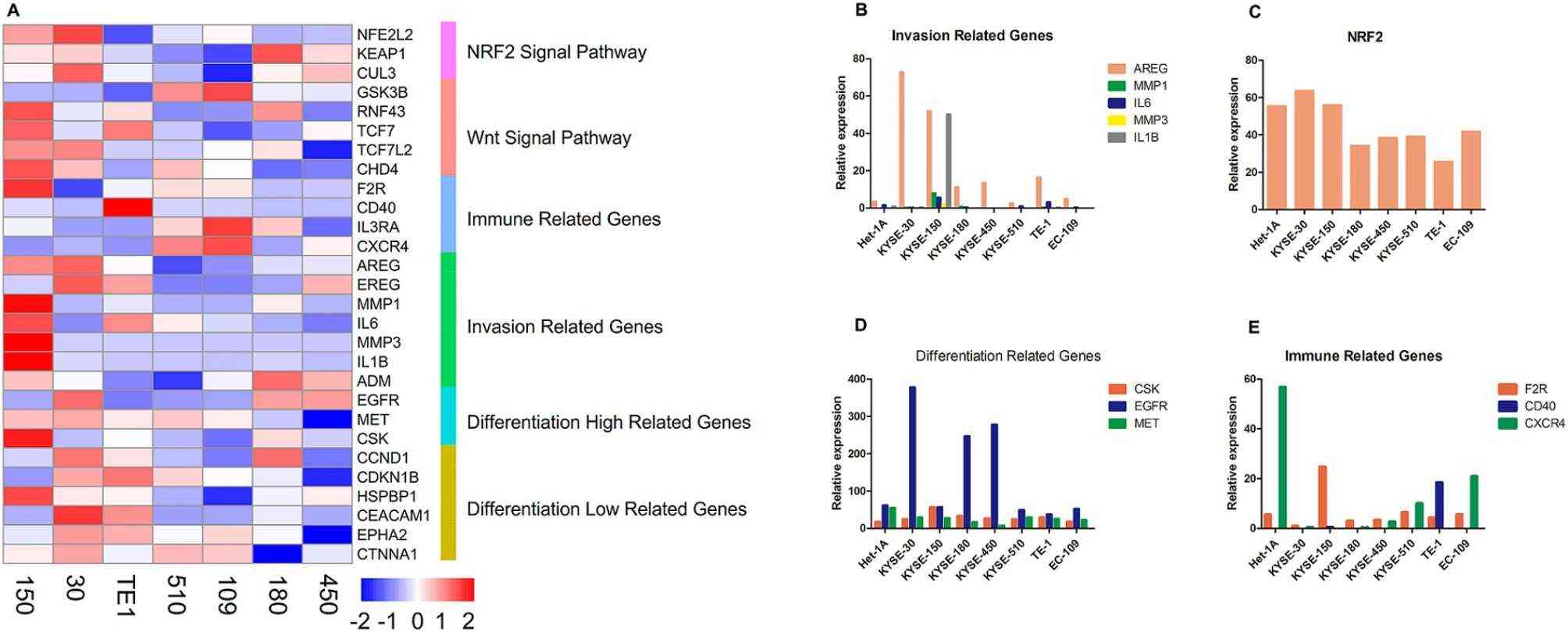 Fig. 2. The expression of oncogenic related genes (Zhang, Chao, et al. 2025).
Fig. 2. The expression of oncogenic related genes (Zhang, Chao, et al. 2025).
Potential 'Anti-Cancer' Effects of Esketamine on Proliferation, Apoptosis, Migration and Invasion in Esophageal Squamous Carcinoma Cells
Esketamine, an N-methyl-D-aspartate receptor antagonist, is commonly used for anesthesia and analgesia clinically. It was reported to negatively regulate cell proliferation, metastasis and apoptosis in cancer cells, including lung cancer and pancreatic cancer. However, its impact on esophageal squamous cell carcinoma (ESCC) malignance and underlying mechanism remain elusive. This study was aimed to investigate the antitumor effects of Esketamine on ESCC in vitro.
The results showed that Esketamine has potential anti-ESCC properties in vitro but subjected to further in vivo and clinical study.
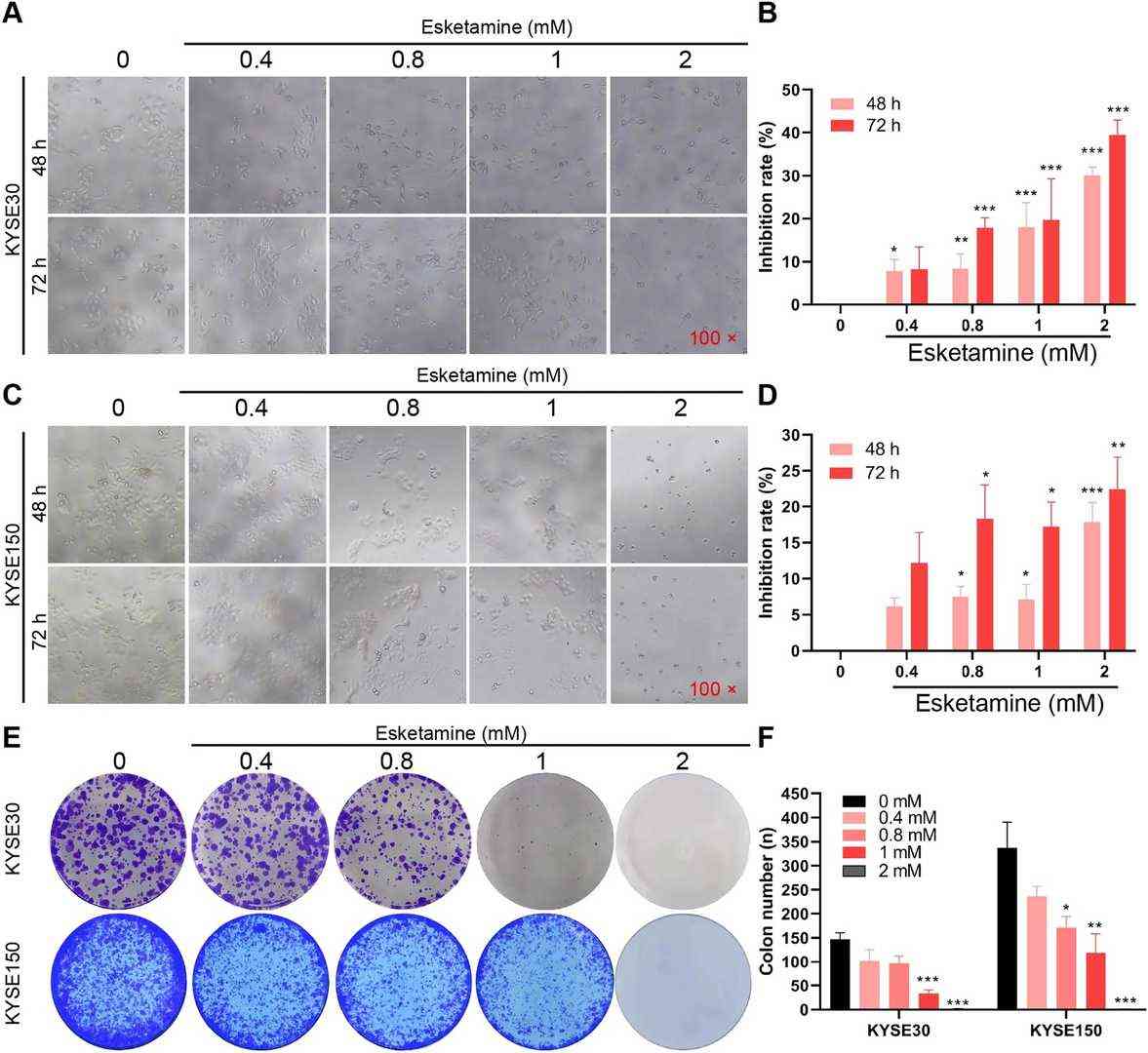 Fig. 3. Esketamine inhibited the viability and proliferation of ESCC cell in vitro (Li, Chao, et al. 2023).
Fig. 3. Esketamine inhibited the viability and proliferation of ESCC cell in vitro (Li, Chao, et al. 2023).
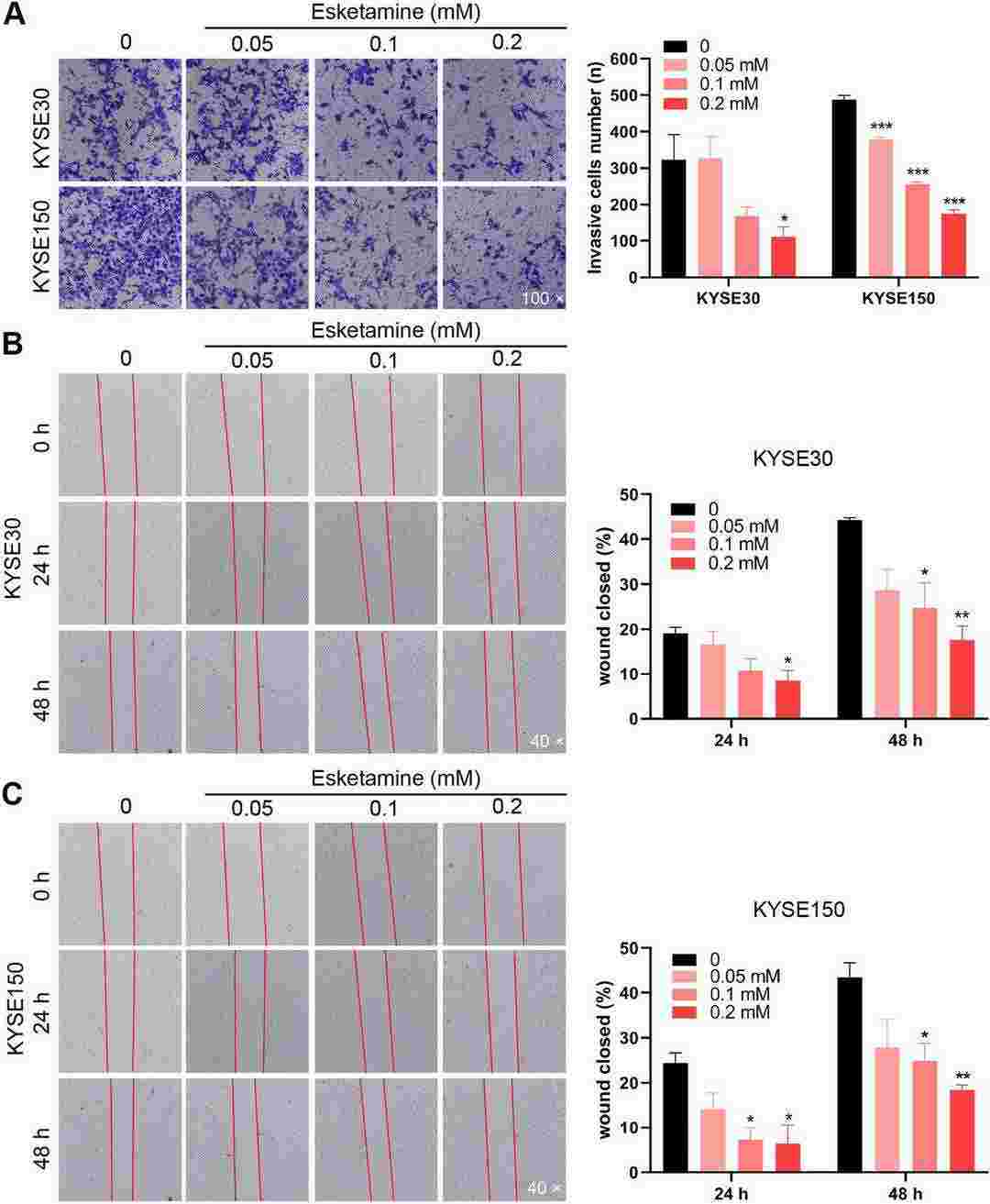 Fig. 4. Esketamine suppressed the migration and invasion ability of ESCC cells (Li, Chao, et al. 2023).
Fig. 4. Esketamine suppressed the migration and invasion ability of ESCC cells (Li, Chao, et al. 2023).
Ask a Question
Write your own review
- You May Also Need
- Adipose Tissue-Derived Stem Cells
- Human Neurons
- Mouse Probe
- Whole Chromosome Painting Probes
- Hepatic Cells
- Renal Cells
- In Vitro ADME Kits
- Tissue Microarray
- Tissue Blocks
- Tissue Sections
- FFPE Cell Pellet
- Probe
- Centromere Probes
- Telomere Probes
- Satellite Enumeration Probes
- Subtelomere Specific Probes
- Bacterial Probes
- ISH/FISH Probes
- Exosome Isolation Kit
- Human Adult Stem Cells
- Mouse Stem Cells
- iPSCs
- Mouse Embryonic Stem Cells
- iPSC Differentiation Kits
- Mesenchymal Stem Cells
- Immortalized Human Cells
- Immortalized Murine Cells
- Cell Immortalization Kit
- Adipose Cells
- Cardiac Cells
- Dermal Cells
- Epidermal Cells
- Peripheral Blood Mononuclear Cells
- Umbilical Cord Cells
- Monkey Primary Cells
- Mouse Primary Cells
- Breast Tumor Cells
- Colorectal Tumor Cells
- Esophageal Tumor Cells
- Lung Tumor Cells
- Leukemia/Lymphoma/Myeloma Cells
- Ovarian Tumor Cells
- Pancreatic Tumor Cells
- Mouse Tumor Cells





