- Specification
- Background
- Scientific Data
- Q & A
- Customer Review
The C106 cell line was established from the tumor tissue of a 78-year-old female patient with moderately well-differentiated adenocarcinoma of the rectum, Dukes' stage A. The cells have epithelial morphology and adherent growth, forming monolayers in culture. They are typically cultured in Iscove's Modified Dulbecco's Medium (IMDM) supplemented with 10% fetal bovine serum (FBS) and 2 mM glutamine, at 37°C and 5% CO₂.
C106 cells harbor mutations in genes commonly altered in colorectal adenocarcinomas, including KRAS and TP53. These mutations contribute to the transformed phenotype and tumorigenicity of the cells. The C106 cell line has been used in various cancer biology studies to investigate cell proliferation, apoptosis, and metastasis. It has also been used for drug screening, especially for drugs targeting KRAS and other related pathways. C106 cells are a useful model for studying the genetic alterations and molecular mechanisms driving colorectal cancer progression. They have been used to explore drug resistance mechanisms, as seen in studies involving KRASG12C and EGFR inhibitors.
Mechanisms of Resistance to Combined KRASG12C and EGFR Inhibition
Despite the therapeutic potential of combined KRASG12C and EGFR inhibition in colorectal cancer, acquired resistance remains a major clinical challenge. Resistance mechanisms are heterogeneous, often involving alterations that sustain ERK signaling. Yaeger et al. characterized these resistance patterns using preclinical models and patient samples, with a focus on identifying dominant mechanisms like KRASG12C amplification.
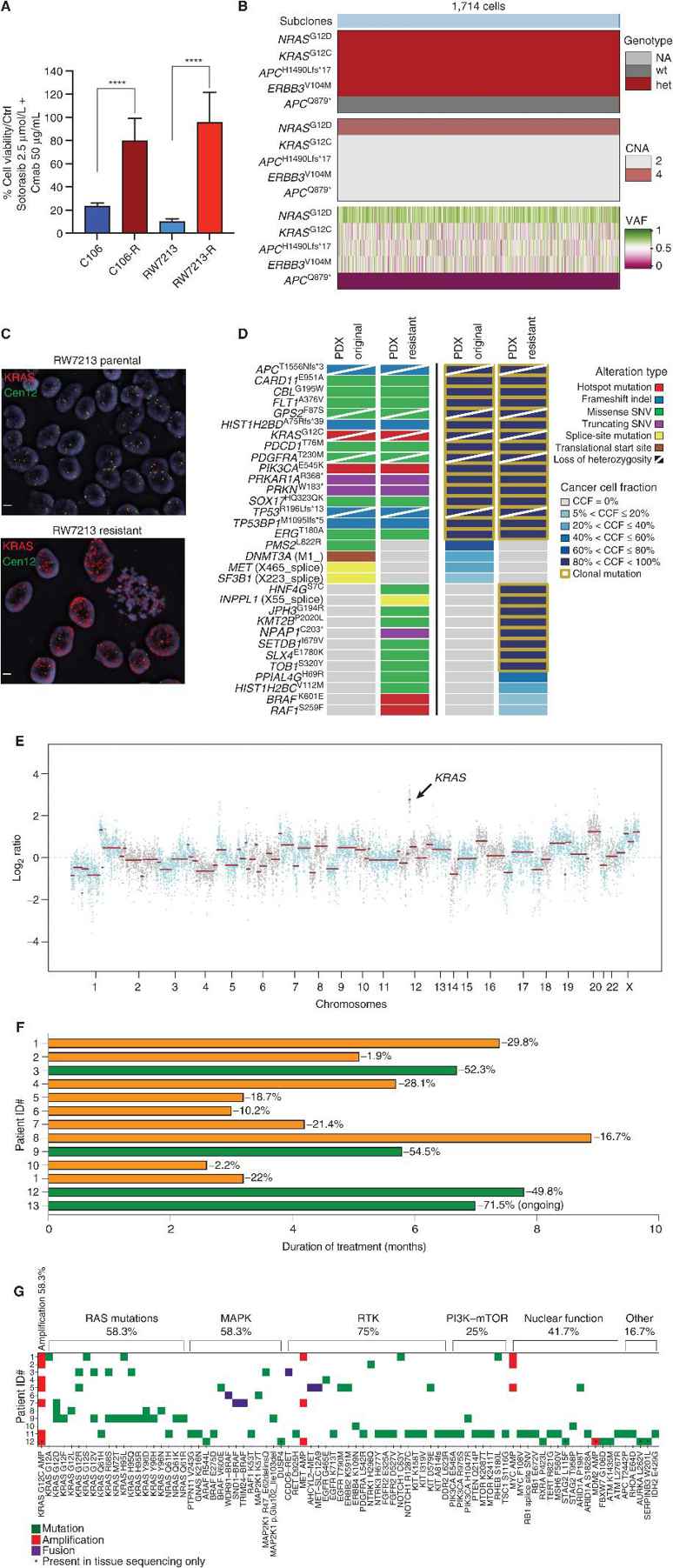
Colorectal cancer cell lines C106 and RW7213 that were sensitive to treatment with sotorasib plus cetuximab were grown in the presence of the drugs until resistant sublines developed (Fig. 1A). RAS-GTP levels in both cell lines were found to be elevated, and pathway suppression was not complete. C106-resistant cells acquired a clonal NRASG12D mutation and late subclonal APCQ879* mutation, while high-level KRASG12C amplification (>20 copies) was found to be the dominant resistance mechanism in RW7213-resistant cells (Fig. 1B–C), as confirmed by FISH analysis. Resistance in a KRASG12C-mutant colorectal PDX model was associated with KRASG12C amplification (VAF 1.00, CCF 100%) after ~10 months on treatment, along with BRAFK601E and RAF1S259F mutations (Fig. 1D–E). This data demonstrates that there can be multiple mechanisms of resistance that allow KRASG12C mutant cells to survive, but KRASG12C amplification is one of the most common alterations at resistance.
Ask a Question
Write your own review
- Adipose Tissue-Derived Stem Cells
- Human Neurons
- Mouse Probe
- Whole Chromosome Painting Probes
- Hepatic Cells
- Renal Cells
- In Vitro ADME Kits
- Tissue Microarray
- Tissue Blocks
- Tissue Sections
- FFPE Cell Pellet
- Probe
- Centromere Probes
- Telomere Probes
- Satellite Enumeration Probes
- Subtelomere Specific Probes
- Bacterial Probes
- ISH/FISH Probes
- Exosome Isolation Kit
- Human Adult Stem Cells
- Mouse Stem Cells
- iPSCs
- Mouse Embryonic Stem Cells
- iPSC Differentiation Kits
- Mesenchymal Stem Cells
- Immortalized Human Cells
- Immortalized Murine Cells
- Cell Immortalization Kit
- Adipose Cells
- Cardiac Cells
- Dermal Cells
- Epidermal Cells
- Peripheral Blood Mononuclear Cells
- Umbilical Cord Cells
- Monkey Primary Cells
- Mouse Primary Cells
- Breast Tumor Cells
- Colorectal Tumor Cells
- Esophageal Tumor Cells
- Lung Tumor Cells
- Leukemia/Lymphoma/Myeloma Cells
- Ovarian Tumor Cells
- Pancreatic Tumor Cells
- Mouse Tumor Cells