Cell Apoptosis Assays
Creative Bioarray provides Cell Apoptosis Assays to all of our customers. The process of programmed cell death, or apoptosis, is generally characterized by distinct morphological characteristics and energy-dependent biochemical mechanisms. Apoptosis is considered a vital component of various processes including normal cell turnover, proper development and functioning of the immune system, hormone-dependent atrophy, embryonic development and chemical-induced cell death. Inappropriate apoptosis is a factor in many human conditions including neurodegenerative diseases, ischemic damage, autoimmune disorders and many types of cancer. The ability to modulate the life or death of a cell is recognized for its immense therapeutic potential. Therefore, research continues to focus on the elucidation and analysis of the cell cycle machinery and signaling pathways that control cell cycle arrest and apoptosis.
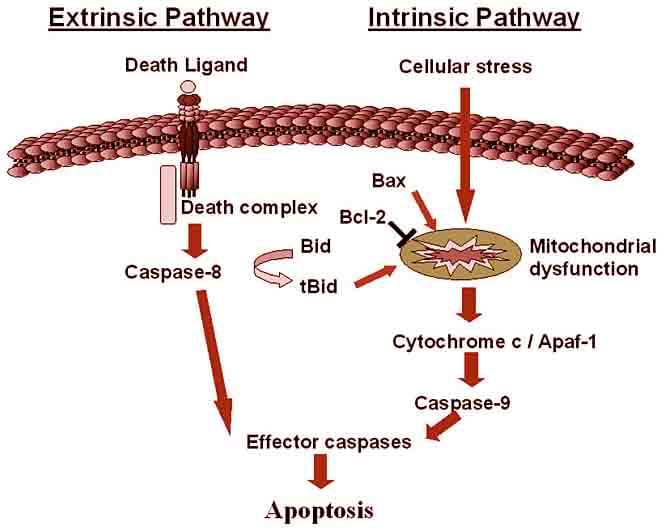 Figure1. Apoptosis pathway
Figure1. Apoptosis pathway
Creative Bioarray offers a wide array of apoptosis assays for measuring multiple components on a choice of assay platforms to accelerate our customer’s research.
- Annexin V binding assays by flow cytometry analysis
Changes in plasma membrane composition and function are detected by the appearance of phosphatidylserine on the plasma membrane, which reacts with Annexin V-fluorochrome conjugates. Combined with staining, this method can distinguish between the early and late apoptotic events.
- Caspase activity assays
Activation of caspase enzymes is a feature of early stage apoptosis. Creative Bioarray offers a wide array of caspase assays including colorimetric/fluorometric substrate-based assays in microtiter plates, detection of cleavage of fluorometric substrate in flow cytometry/microscopy or by microtiter plates analysis, Western blot analysis of pro-and active caspase, flow cytometry microscopy/microplate spectrophotometry analysis with antibodies specifically recognizing the active form of caspases.
- Mitochondrial membrane potential assays
Mitochondria play an important role in this process that can be recognized by translocation of apoptogenic factors, such as Bax and cytochrome c, in and out of mitochondria.
- DNA fragmentation and morphology
Apoptotic cells are recognized on the basis of their reduced DNA content and morphological changes that include nuclear condensation and which can be detected by flow cytometry (sub-G1 DNA content), Trypan Blue, or Hoechst staining. Our assays provide quick and reliable detection methods for studying in situ apoptotic DNA fragmentation.
- Other assays available
Other assays are available based on Nuclear condensation, Non-caspase proteases (cathepsins and calpain) activation, Caspase substrate (PARP) cleavage, and Cleavage of anti-apoptotic Bcl-2 family proteins.
Creative Bioarray Advantages
- Various cell/tissue types we can handle: live cells, cell extracts, tissue extracts or fixed cells
- Analysis techniques available: Flow cytometry, Western blot, Microscopy and Colorimetric/fluorometric substrate-based assays
- Superior performance-highly sensitive assays to detect different phase of apoptosis: Prophase, Pro&metaphase, Anaphase
- Diverse selection-large number of assays to detect different parameters
- Fast turnaround time
Result Example
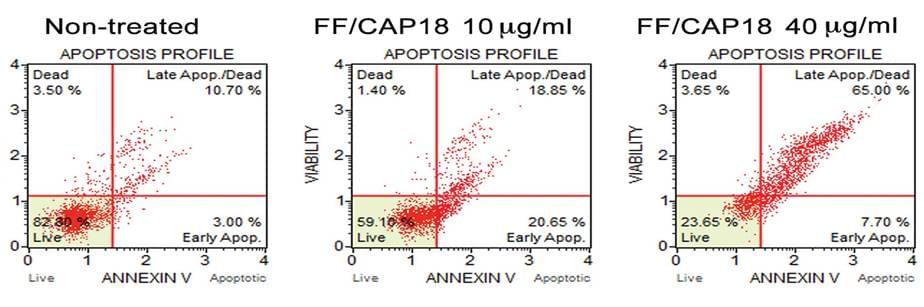 Figure 2 Annexin V binding assays -Detection of apoptosis of HCT116 cells
Figure 2 Annexin V binding assays -Detection of apoptosis of HCT116 cells
Quotations and Ordering
Our customer service representatives are available 24hr a day! We thank you for choosing Creative Bioarray services!
If you want to perform the analysis in house, Creative Bioarray also provides Cell Apoptosis analysis kit to assist with your research.
References
- Elmore, S. Apoptosis: a review of programmed cell death. Toxicologic Pathology, 2007, 35(4): 495-516.
- Kanduc. D.; et al. Cell death: apoptosis versus necrosis. International Journal of Oncology, 2002, 21: 165-170.
Explore Other Options