Cell-based LNP Evaluation
LNPs (Lipid Nanoparticles) are widely used in the field of gene therapy and drug delivery, particularly for the delivery of nucleic acids such as mRNA or siRNA into cells. Creative Bioarray offers a range of assays specifically designed to evaluate the behavior and effects of LNPs on cells. These assays are aimed at providing valuable insights into LNP cellular uptake, intracellular trafficking, gene expression, cytotoxicity, and other relevant parameters.
1. LNP uptake assessment: Cell-based imaging or fluorescence detection commonly employed to assess the cellular uptake of LNPs. Visualization techniques such as fluorescent labeling enable tracking of LNPs within the cells.
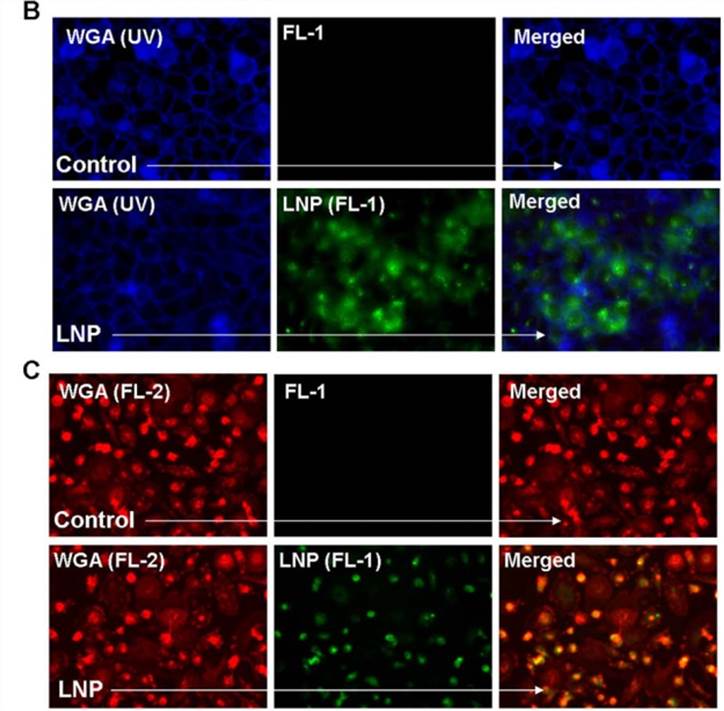 Figure 1. Lipid nanoparticle (LNP) uptake in various cell types. (B) The membrane stain wheat germ agglutinin (WGA) visualized in blue was used with fluorescent microscopy in order to visualize the uptake of LNP-FITC (green) after a 16 hr incubation with HeLa cells. (C) WGA visualized in red was used to observe LNP-FITC (green) uptake in primary macrophages.[1]
Figure 1. Lipid nanoparticle (LNP) uptake in various cell types. (B) The membrane stain wheat germ agglutinin (WGA) visualized in blue was used with fluorescent microscopy in order to visualize the uptake of LNP-FITC (green) after a 16 hr incubation with HeLa cells. (C) WGA visualized in red was used to observe LNP-FITC (green) uptake in primary macrophages.[1]
2. LNP intracellular trafficking analysis: Investigate the fate of LNPs after cellular uptake. Determine whether the LNPs are efficiently released within the cytoplasm or reach specific organelles of interest. Techniques such as colocalization studies with organelle-specific markers or biochemical fractionation can provide insights into intracellular trafficking pathways.
- Membrane fusion detection: The dye dilution method is a technique used to detect and study membrane fusion events involving LNPs and cell membranes.
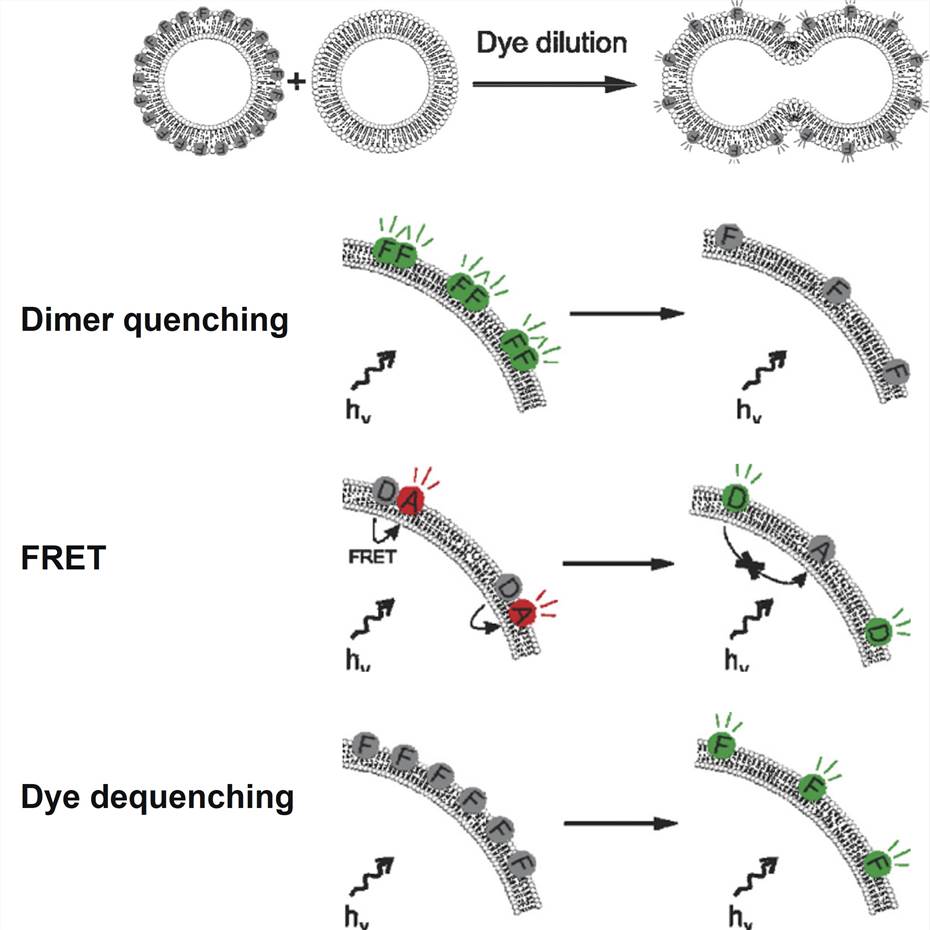 Figure 2. Successful lipid—lipid fusion leads to a dilution of the incorporated fluorophores over a larger surface area. Dye dilution can be detected in a number of ways: dimer quenching, resulting in loss of fluorescence; FRET, resulting in a shift in fluorescence ratio of two fluorophores; dye dequenching, resulting in an increase in fluorescence due to dequenching of a self-quenching fluorophore.[2]
Figure 2. Successful lipid—lipid fusion leads to a dilution of the incorporated fluorophores over a larger surface area. Dye dilution can be detected in a number of ways: dimer quenching, resulting in loss of fluorescence; FRET, resulting in a shift in fluorescence ratio of two fluorophores; dye dequenching, resulting in an increase in fluorescence due to dequenching of a self-quenching fluorophore.[2]
- Co-localization imaging: Fluorescence co-localization imaging of nucleic acid fluorescent markers and endosomal markers.
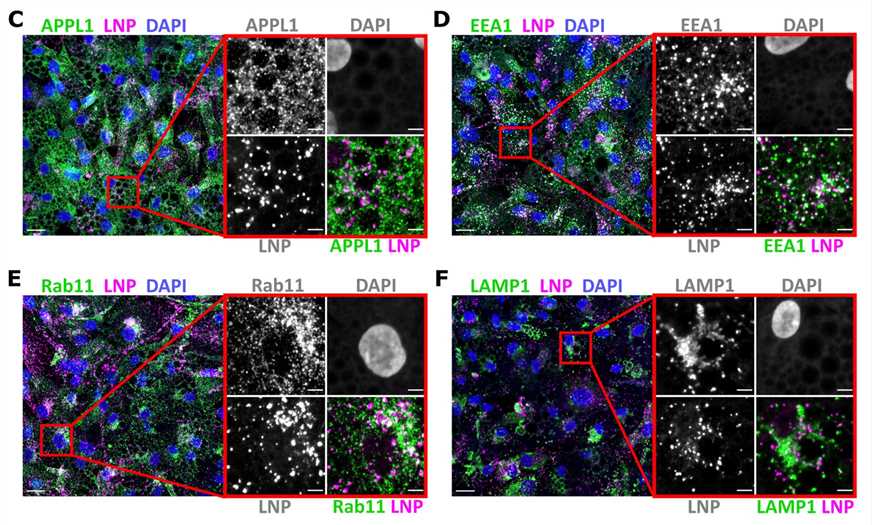 Figure 3. Representative images of human primary adipocytes incubated with L608 LNP-mRNA for 2 h and immunostained with antibodies against endosomal markers (in green) as follows: APPL1 (C), EEA1 (D), Rab11 (E), and LAMP1 (F). Exogenous mRNA was detected by smFISH (labeled as LNP) and nuclei by DAPI. The magnified area is presented with split and merged color images. Scale bars are 20 μm in the overview and 5 μm in the magnified images.[3]
Figure 3. Representative images of human primary adipocytes incubated with L608 LNP-mRNA for 2 h and immunostained with antibodies against endosomal markers (in green) as follows: APPL1 (C), EEA1 (D), Rab11 (E), and LAMP1 (F). Exogenous mRNA was detected by smFISH (labeled as LNP) and nuclei by DAPI. The magnified area is presented with split and merged color images. Scale bars are 20 μm in the overview and 5 μm in the magnified images.[3]
- Efficient release of LNPs:LNPs typically carry labeled cargo, such as nucleic acids tagged with fluorescence, e.g., Cy5. Upon cellular uptake, LNPs accumulate in endosomes, resulting in a distinctive punctate phenotype. This fluorescence-based approach enables visualization and assessment of efficient LNP cargo release within the cellular environment.
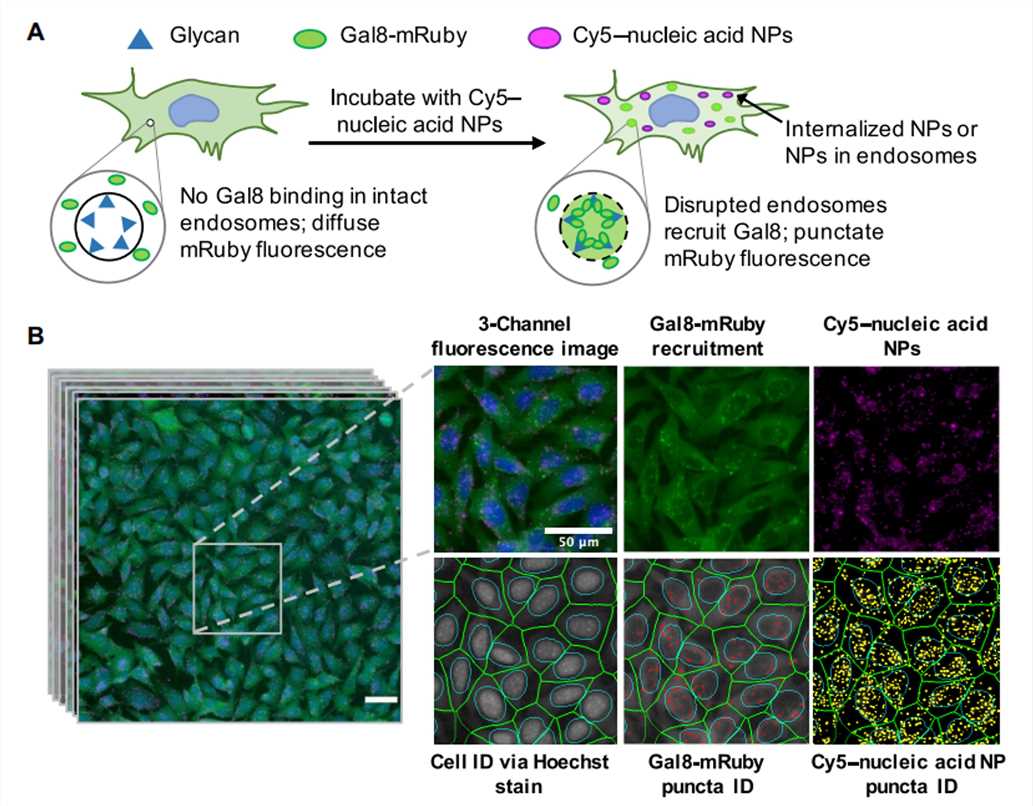 Figure 4. Assay overview. Cells genetically encoding a Gal8-mRuby fusion fluorescence protein exhibited diffuse cytosolic mRuby signal in the absence of endosomal disruption. Endosomal disruption caused by NPs carrying Cy5-labeled nucleic acid NPs allows Gal8-mRuby to bind to intra-endosomal glycans, resulting in punctate fluorescent spots.[4]
Figure 4. Assay overview. Cells genetically encoding a Gal8-mRuby fusion fluorescence protein exhibited diffuse cytosolic mRuby signal in the absence of endosomal disruption. Endosomal disruption caused by NPs carrying Cy5-labeled nucleic acid NPs allows Gal8-mRuby to bind to intra-endosomal glycans, resulting in punctate fluorescent spots.[4]
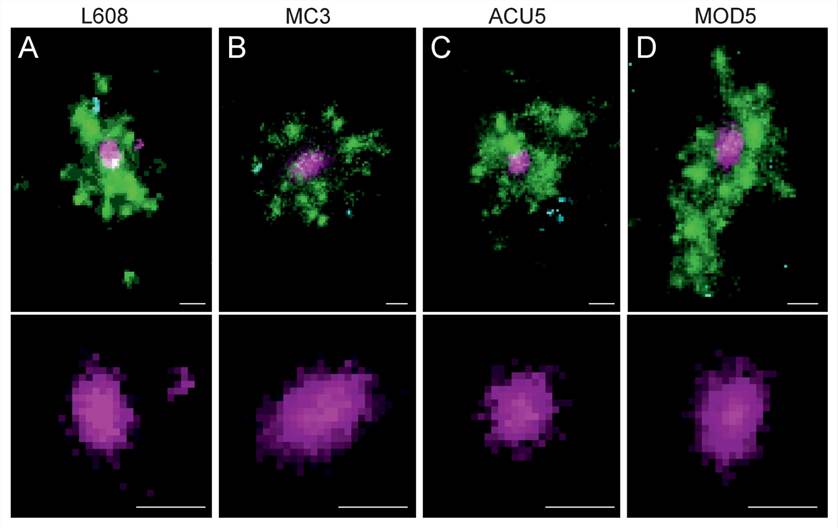 Figure 5. Multicolor SMLM detects and visualizes singular LNP-mRNA in endosomal compartments with nanometer resolution. (A–D) Exemplary images of endosomes containing a single, clearly detectable LNP-Cy5-mRNA (magenta) together with transferrin (green) and EGF (cyan) cargo for each imaged LNP formulation (top row).[3]
Figure 5. Multicolor SMLM detects and visualizes singular LNP-mRNA in endosomal compartments with nanometer resolution. (A–D) Exemplary images of endosomes containing a single, clearly detectable LNP-Cy5-mRNA (magenta) together with transferrin (green) and EGF (cyan) cargo for each imaged LNP formulation (top row).[3]
3. Gene expression: Evaluate LNP-mediated gene expression efficacy for nucleic acid cargo (e.g., mRNA) through quantification of the target gene's expression using qPCR, western blotting, or reporter assays.
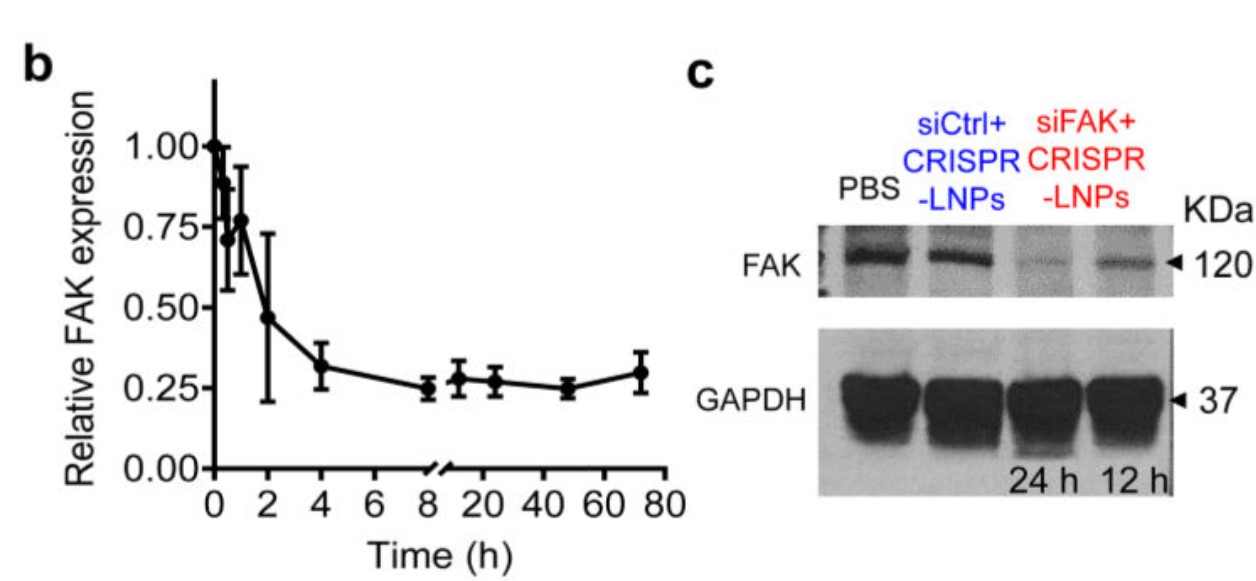 Figure 6. siRNA-mediated gene silencing successfully inhibited FAK expression. b. RT-qPCR quantification of time-dependent FAK expression; c. Western blot analysis of FAK expression.[5]
Figure 6. siRNA-mediated gene silencing successfully inhibited FAK expression. b. RT-qPCR quantification of time-dependent FAK expression; c. Western blot analysis of FAK expression.[5]
4. Cytotoxicity: Determine the potential cytotoxic effects of LNPs on the treated cells.
- Cell viability/proliferation assays
MTT assay
CCK8 assay
alamarBlue assay - Cell membrane integrity assays
LDH assay
Glycerol-3-Phosphate Dehydrogenase (G3PDH) assay
Adenosine kinase (AK) luminescence assay
Propidium Iodide (PI) labeling - Cellular metabolism assay
GSH assay - Mitochondrial function assays
Quantitative detection of ATP
ROS detection
5. Additional assays: Depending on the specific research objectives, other assays may be included in the LNP evaluation assays. These can include functional assays to evaluate the biological impact of LNP-mediated delivery, immunogenicity assessments, or any other relevant experimental endpoints.
It's important to note that the exact details of the LNP evaluation assays may vary depending on the specific research question and objectives. For personalized guidance, please contact our specialists to discuss your specific needs.
References
- Kovochich, Michael et al. "Activation of latent HIV using drug-loaded nanoparticles." PloS one vol. 6,4 e18270. 5 Apr. 2011, doi:10.1371/journal.pone.0018270
- Martens, Thomas F., et al. "Intracellular delivery of nanomaterials: how to catch endosomal escape in the act." Nano Today 9.3 (2014): 344-364. DOI:10.1016/j.nantod.2014.04.011.
- Paramasivam, Prasath et al. "Endosomal escape of delivered mRNA from endosomal recycling tubules visualized at the nanoscale." The Journal of cell biology vol. 221,2 (2022): e202110137. doi:10.1083/jcb.202110137
- Rui, Yuan et al. "High-throughput and high-content bioassay enables tuning of polyester nanoparticles for cellular uptake, endosomal escape, and systemic in vivo delivery of mRNA." Science advances vol. 8,1 (2022): eabk2855. doi:10.1126/sciadv.abk2855
- Zhang, Di et al. "Enhancing CRISPR/Cas gene editing through modulating cellular mechanical properties for cancer therapy." Nature nanotechnology vol. 17,7 (2022): 777-787. doi:10.1038/s41565-022-01122-3
Explore Other Options