Comparative Genomic Hybridization Protocol
GUIDELINE
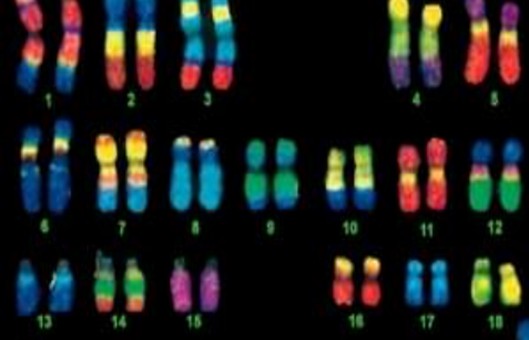
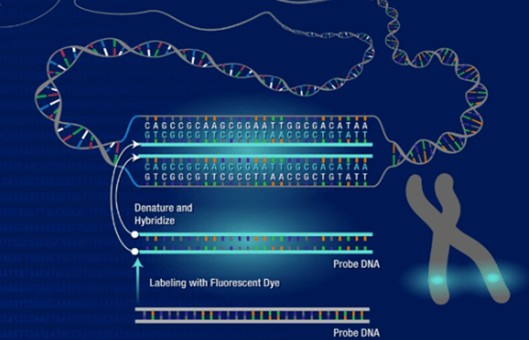
Comparative genomic hybridization (CGH) allows genome-wide assessment of changes in relative copy number of DNA sequences, using tumor-cell DNA extracted from clinical samples or cell lines as a probe. In CGH, amplifications, gains, and deletions of genes are detected using a modified fluorescence in situ hybridization (FISH) reaction.
METHODS
Prepare probe mix
- Combine the following in a 1.5-ml microcentrifuge tube, 10 µl 20 µg/ml fluorescein-labeled test probe DNA (200 ng final), 10 µl 20 µg/ml Texas red-labeled reference probe DNA (200 ng final), and 20 µl 1 µg/µl human Cot-1 DNA.
- Add 1/10 vol of 3 M sodium acetate, pH 5.2, mix, then precipitate DNA by adding 2.5 vol of 100% ethanol and mixing again. Microcentrifuge 30 min at maximum speed, 4°C.
- Decant the supernatant and blot the excess liquid from the inside of the tube, taking care to avoid blotting the pellet. Allow the pellet to air dry for 10 min.
- Add 10 µl master hybridization solution and resuspend the pellet gently with the pipet tip. Vortex, then microcentrifuge 10 sec at maximum speed to bring the liquid to the bottom of the tube.
Denature target metaphase slides
- Outline the area on the back of the slide containing metaphase chromosomes using a diamond pen.
- Prewarm denaturation solution to 73°C in a Coplin jar placed in a 75°C water bath. Prewarm slides<2 min on a 37°C slide warmer and place them (no more than three to four at a time) in the denaturation solution. Incubate for 2.5 to 10 min at 73°C.
- Dehydrate slides by immersing them for 2 min successively in Coplin jars containing 70%, 85%, and 100% ethanol at room temperature. Remove from ethanol and air dry in a vertical position to minimize spotting.
- Immerse slides in proteinase K solution for 5 to 7 min at room temperature. Repeat the previous step.
Denature probe and hybridize to slides
- Place slides on a 37°C slide warmer. Denature probe mix from step 4 by heating for 5 min at 70°C, then (with slides still on warmer) immediately add 10 µl probe mix to the area of each slide containing the metaphases. Cover with an 18-mm2 coverslip, removing any air bubbles by gently lowering the coverslip onto the slide at an angle using fine-tipped forceps. Seal coverslip edges with rubber cement.
- Incubate in a moist chamber for 2 to 3 days at 37°C.
Wash hybridized slides
- Peel off rubber cement and gently remove coverslips. Wash slides by immersing successively for 10 min each in three changes of 45°C hybridization wash buffer prewarmed to 45°C.
- Wash slides by immersing for 10 min in 45°C 2×SSC, then 10 min in room temperature 2×SSC.
- Wash slides by immersing successively for 5 min each in two changes of room temperature PN buffer, then successively for 5 min each in two changes of room temperature distilled water. Air dry.
Counterstain chromosomal DNA and view slides
- Apply 8 µl DAPI in antifade and cover with a 22-mm2 coverslip.
- Examine and analyze slides.
Creative Bioarray Relevant Recommendations
- CGH is one of the most widely used methods to detect the amplification and large-scale acquisition or loss of genetic material because it can achieve a genome-wide screening imbalance in a single experiment. We establish a platform for traditional CGH analysis and optional micro-CGH analysis based on FISH services.
- We also provide Array CGH Services, a high-resolution karyotype analysis solution, for the detection of unbalanced structural and numerical chromosomal alterations with high-throughput capabilities. In addition, a wide range of Karyotyping Services (Traditional Karyotyping-G Banded, M-FISH, Molecular Karyotyping, etc.) is provided to meet our customer's different needs.
NOTES
- The amounts of test and reference probe DNA can be increased in subsequent experiments if the resulting hybridization signals appear too dim.
- The quality of CGH results varies with the metaphase preparation. Metaphase slides that work well for FISH may not be adequate for CGH. It is best to make large batches of slides and to optimize the denaturation conditions used in the following steps for each batch. CGH quality with a particular batch of metaphase slides may deteriorate over time.
- Proteinase K digests nuclear protein, cytoplasm, and debris, reducing background and increasing probe penetration. Although proteinase K digestion may yield brighter and smoother hybridizations, this treatment may diminish the quality of chromosome banding and result in chromosomes that appear indistinct and fuzzy. If proteinase K treatment is necessary, the time of treatment must be optimized (between 5 and 7 min) so that chromosome morphology is maintained and identification by banding pattern remains feasible.