Cardiomyocyte Differentiation Service from iPSC
iPSCs have great replicative ability and demonstrated potential to form functional cardiomyocytes (CMs). These CMs stand for a promising source for cell replacement therapy to treat heart disease, and they may act as a potent tool for drug discovery and disease modeling. Until now, a variety of approaches have been developed for producing iPSC-derived CMs and many are available in Creative Bioarray.
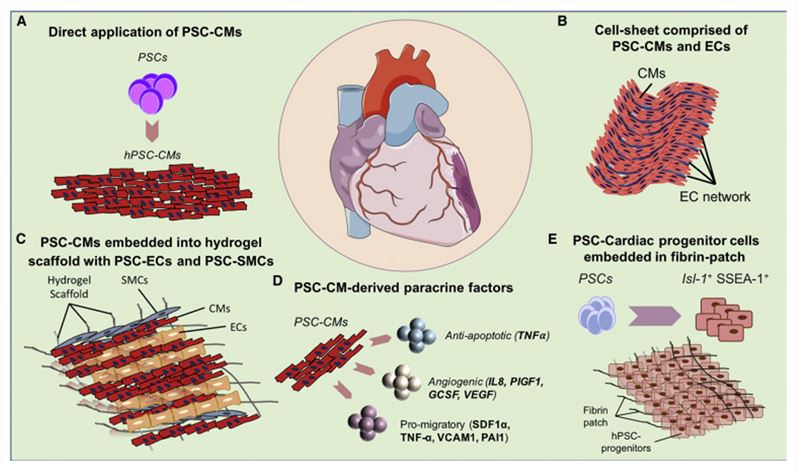 Figure 1. Cardiac Cell Therapy Using PSC-CMs.
Figure 1. Cardiac Cell Therapy Using PSC-CMs.
Creative Bioarray provides top quality service for directed-differentiation of iPSCs into cardiomyocytes using our proprietary induction protocol and reagents. We deliver ready-to-use, functional cardiomyocytes that are desirable in vitro models for high content toxicity and drug screening, and cell regeneration research. They are a feasible and physiologically relevant alternative to embryonic stem cell, primary cells, and animal models.
Workflow:
- Recovery, Expansion and Validation of iPSCs/ESCs
- Cardiomyocyte Differentiation
- Characterization of differentiated cells by immunocytochemical (ICC) staining: 2 biomarkers (cTNT; SMA)
Deliverables:
1. Lineage-committed cardiomyocytes differentiated from iPSCs.
2. Report containing results and data:
- Phase contrast images of parental iPSC lines after expansion (milestone 1); and differentiated cardiomyocytes
- Videos of beating cardiomyocytes (before cryopreservation)
- Antibody staining images of cardiomyocytes using anti-TNT anti-SMA antibodies
- Freeze-thaw viability and mycoplasma testing
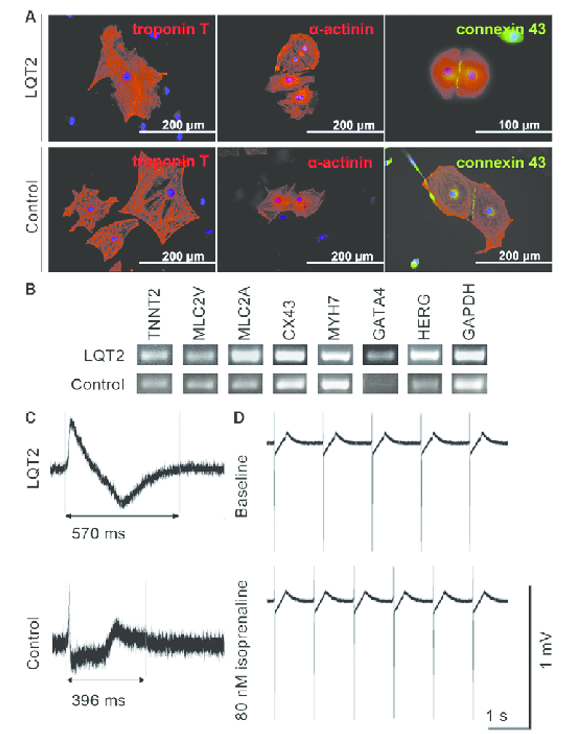 Figure 2. The expression of cardiac markers in iPSC-derived cardiomyocytes and the electrical properties of the cells.
Figure 2. The expression of cardiac markers in iPSC-derived cardiomyocytes and the electrical properties of the cells.
Creative Bioarray is an experienced and outstanding provider of Cardiomyocyte Differentiation service as well as differentiation kit. We are dedicated to providing quality data and comprehensive service for your scientific research, and we are pleased to use our extensive experience and advanced platform to offer the best service to satisfy each demand from our customers.
If you have any special need in Cardiomyocyte Differentiation service, do not hesitate to contact us for this special service. Please let us know what you need and we will accommodate you. We look forward to working with you in the future.
Application of iPSC-Derived Cardiomyocytes
Human induced pluripotent stem cell-derived cardiomyocytes (iPSC-CMs) have emerged as promising tools for cardiac research. Theoretically, iPSC-CMs provide an accessible source of human cardiomyocytes without the ethical and practical concerns of using human cardiac tissue or cells. From the experimental point of view, iPSC-CMs also resolve problems related to comparisons between species, thus facilitating translation between basic research and clinical science. Furthermore, since iPSC-CMs retain the genetic identity of the individual donor, they enable generation of patient- and disease-specific cells that can be employed in procedures of personalized medicine.
Cardiac disease modeling
In vitro disease modeling allows for investigation of pathological mechanisms as well as evaluation of therapeutic options (e.g., pharmaceutical efficacy, etc.). Distinctions between healthy and diseased cardiomyocytes are typically made using cardiac functional assessments in addition to structural (e.g., organelle morphology, sarcomere structure) and molecular (e.g., aberrant signaling pathways) evaluations. To date, arrythmias, cardiomyopathies, cardiometabolic disorders, and multifaceted cardiac diseases with yet-to-be-defined genetics have been modeled in vitro using iPSC-CMs.
Cardiotoxicity screening
Cardiotoxicity and arrhythmia induction, such as life-threatening torsade de pointes (TdP), are major causes of preclinical and clinical drug failure and withdrawal from the market. In 2013, the Comprehensive in Vitro Proarrhythmia Assay (CiPA) initiative was founded to overcome the low specificity of the preclinical studies and clinical trials at that time. One of the novel components is testing the effect of a drug in vitro in iPSC-CMs.
3D heart-on-a-chip models are also being tested, holding promise for even better prediction of cardiotoxic and pro-arrhythmic drug effects as they better recapitulated the clinical effects compared to 2D iPSC-CMs models as they present occasionally with arrhythmias that are not reported in adult cardiomyocytes.
Gene therapy testing
Inherited cardiac arrhythmia iPSC-CMs models have also been used to test novel gene therapies, acting straight on the nucleic acid molecular/genetic level.
Scientific Data
Differentiation and characterization of cardiomyocytes
A two-step procedure was used to compare two monolayer-based serum-free differentiation protocols: mesoderm induction through activation of the Wnt/β-catenin pathway using medium supplemented with a GSK3 inhibitor – CHIR99021, followed by cardiac-fate determination through inhibition of the canonical Wnt pathway using media supplemented with IWP2 (protocol A) and Wnt-C59 (protocol B) (Fig. 1B).
All generated iPSC-CMs showed expression of all cardiac protein markers (cACT, cTNNI and Nkx2.5) tested in ICC staining for protocol A and B (Fig. 2A, B). iPSC-CMs differentiated using protocol A showed less organization of cTNNI, where generated cells showed cTNNI expression mainly around the nucleus and not presenting cytoskeletal structures visible in the cells obtained with protocol B (Fig. 2B). Average differentiation efficiency calculated from ICC staining of cACT was similar for iPSC-CMs generated using both protocols (Fig. 2A).
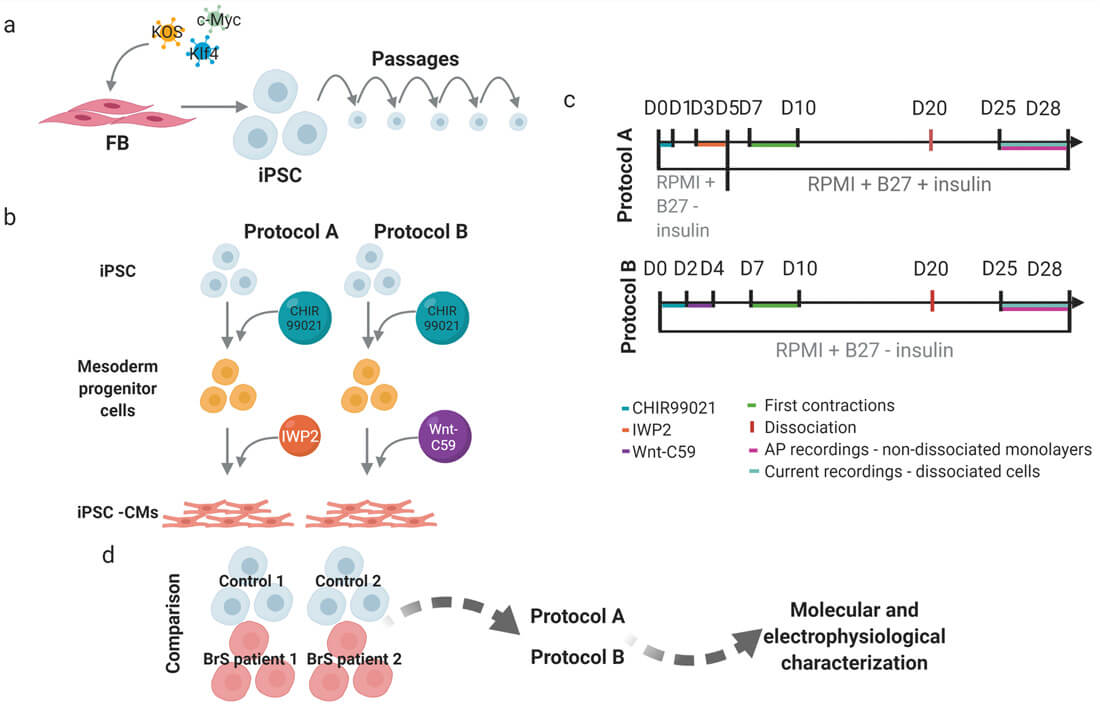 Fig. 1. Schematic presentation of the workflow and experimental design (Aleksandra Nijak, et al., 2022).
Fig. 1. Schematic presentation of the workflow and experimental design (Aleksandra Nijak, et al., 2022).
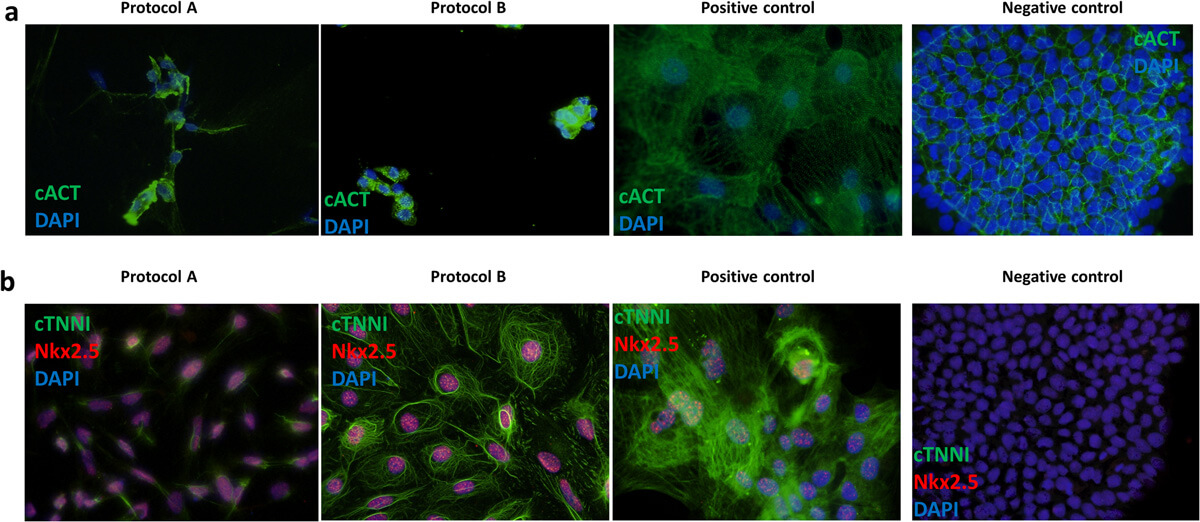 Fig. 2. Representative immunofluorescence staining of cardiac markers (Aleksandra Nijak, et al., 2022).
Fig. 2. Representative immunofluorescence staining of cardiac markers (Aleksandra Nijak, et al., 2022).
Transcript levels of selected cardiac ion channel and structural genes in generated iPSC-CMs as fold expression compared to iPSCs
To select the best performing differentiation protocol, one iPSC clone from four individuals (Control 1 and 2, BrS patient 1 and 2) was used and each differentiated in four or six replicates (one well on a six-well plate=one replicate) using protocol A and B (Fig. 1D).
Evaluating the mRNA levels of selected cardiac markers in iPSC-CMs for both protocols (except for control 2 for which insufficient RNA concentration was obtained), the results showed significant differences in the expression levels for five of the markers (GJA1, KCNJ2, MYH7, RYR2, TNNI3) between the used protocols (Fig. 3). Compared with left ventricular (LV) cardiac tissue, the expression of KCND3, KCNQ1, RYR2, TNNI3 and TNNT2 was significantly lower in iPSC-CMs obtained by both protocols (Fig. 3). Additionally, cells generated with protocol A showed significantly lower relative expression of KCNH2, KCNJ8, MYH6 and MYH7, as well as higher relative expression of KCNJ2 and GJA1, while cells generated with protocol B showed significantly higher relative expression of MYH6 compared with the reference tissue. Although both protocol A and B show differences in expression compared to LV tissue, protocol B showed more similar expression levels of each of the tested genes in comparison to LV tissue.
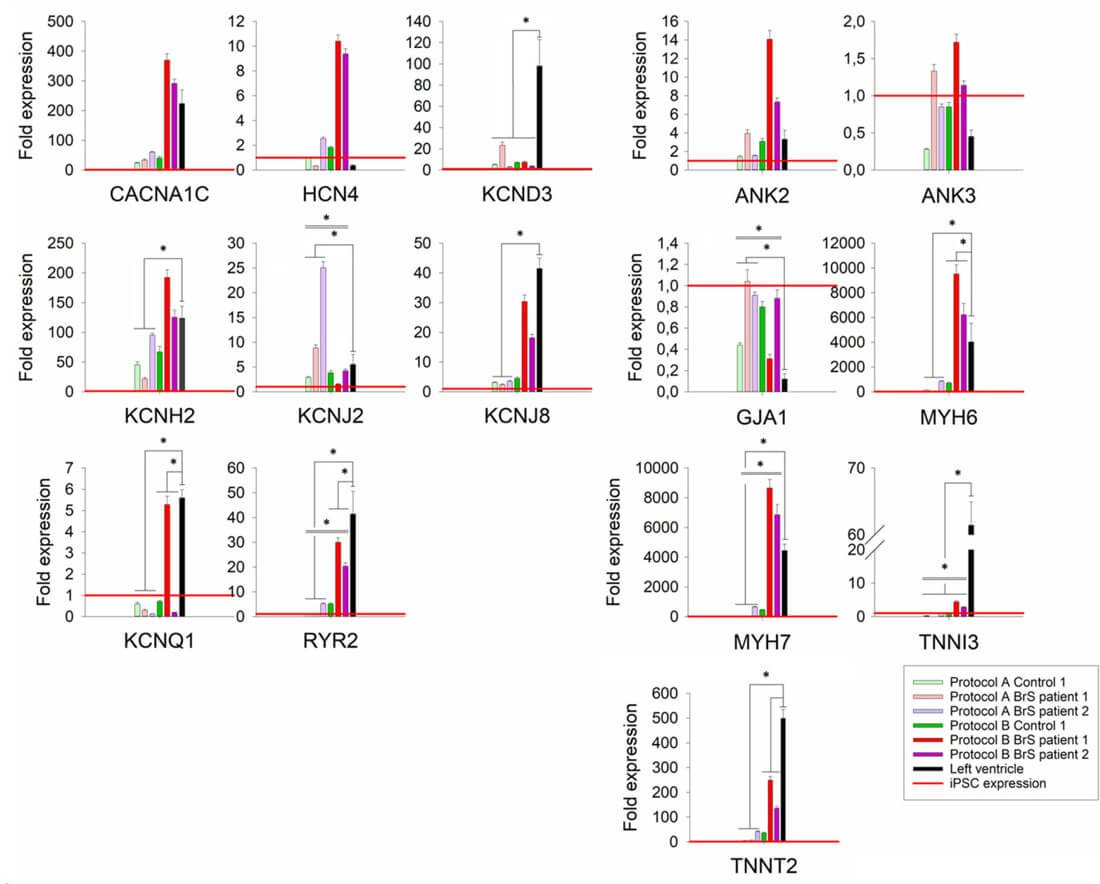 Fig. 3. Relative expression levels of tested cardiac markers for cells differentiated with protocols A and B (Aleksandra Nijak, et al., 2022).
Fig. 3. Relative expression levels of tested cardiac markers for cells differentiated with protocols A and B (Aleksandra Nijak, et al., 2022).
Patch-clamping experiment results for iPSC-CMs in the comparison of protocol A and B
An important feature of the created iPSC-CMs is proper survival after dissociation to allow single-cell patch-clamp recordings, especially required for characterization of specific ionic currents. For AP characterization, we performed AP recordings on a non-dissociated monolayer, as these reflect a more physiologically relevant condition. Based on the numbers of recordings per cell line (average 6.7 for protocol A and 10.7 for protocol B; Table 1), the cells generated with protocol B were more approachable (flatter and more elongated) for patch-clamp experiments than those obtained with protocol A.
Table 1. Action potential properties for iPSC-CMs generated in the comparison experiment (Aleksandra Nijak, et al., 2022).
| Protocol A | Protocol B | |||||
| Cell line | Control 2 | BrS patient 1 | BrS patient 2 | Control 2 | BrS patient 1 | BrS patient 2 |
| Number of AP recordings | 4 | 8 | 8 | 10 | 9 | 13 |
| Number of analysed AP recordings | 0 * | 0 */† | 1 | 0 */‡ | 4 | 2 |
| APD50 [ms] | - | - | 422.08 | - | 351.51± 24.84 | 273.7± 40.83 |
| APD90 [ms] | - | - | 501.8 | - | 509.95± 27.91 | 422.3± 10.9 |
| APA [mV] | - | - | 114.5 | - | 90.8± 5.1 | 86.7± 17.3 |
| RMP [mV] | - | - | -59.3 | - | -59.6± 3.1 | -58.3± 5.4 |
| BPM | - | - | 8 | - | 17± 1.12 | 27± 3 |
FAQ
1. What do I need to supply for this service?
Cells to be differentiated: Ideally 2-3 vials of frozen stocks on dry ice or one T25 flask of live cells per sample. Creative Bioarray also provides a wide collection of iPSCs for the customers to choose from.
2. I don't have my own iPSCs, can you help me to order an iPSC line?
Creative Bioarray provides a wide collection of iPSCs from normal or diseased donors. In addition, we help our customer to develop their own iPSC models with our Reprogramming and Genome Editing services.
3. How long is the turnaround time?
It takes 2-3 months depending on the recovery of the cells provided by the customer, cell type to be differentiated, and the scale of differentiation.
4. Do you provide functional tests or other characterization services for my iPSC-derived cells?
Yes, please contact us to learn more about characterization and functional assays.
5. What are the deliverables?
Cryovials of differentiated cells or live culture for cell types not suitable for cryopreservation.
Reports and data.
References
- Nijak, Aleksandra, et al. Morpho-functional comparison of differentiation protocols to create iPSC-derived cardiomyocytes. Biology Open, 11.2 (2022): bio059016.
- Lian, Xiaojun, et al. Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/β-catenin signaling under fully defined conditions. Nature protocols, 8.1 (2013): 162-175.
- Burridge, Paul W., et al. Chemically defined generation of human cardiomyocytes. Nature methods, 11.8 (2014): 855-860.
- Van de Sande, Dieter V., et al. The resting membrane potential of hSC-CM in a syncytium is more hyperpolarised than that of isolated cells. Channels, 15.1 (2021): 239-252.

